Analysis
1 Packages
library(tidyverse)
library(metan)
library(rio)
library(ggrepel)
library(R2OpenBUGS)
library(boa)
library(ggformula)To perform the bayesian analysis, the Open BUGS software need to be installed. Follow these instructions to install it.
2 Data
data_cv <-
import("http://bit.ly/data_cvs") %>%
select(GY:NGS)
# Long format
data_cv_long <-
data_cv %>%
pivot_longer(everything(),
names_to = "var",
values_to = "cv") %>%
remove_rows_na()
# samples per variable
data_cv_long %>% n_by(var)
# create a list of variables with no missing values
df <- lapply(data_cv, remove_rows_na)3 Bayesian
3.1 Function
bayes <- function(df){
linemodel <- function(){
for (i in 1:64) # change the number of samples for each variable
{
y[i] ~ dgamma(r, mu)
}
r ~ dunif(0,5)
mu ~ dunif(0,5)
}
################ Especificando os dados
linedata <- list(y = df[[1]])
###################### Specification innitial values
lineinits <- function(){list(r = 0.5, mu = 1) }
#Specification the parameters
parameters <- c("r","mu")
############# Execution function analysis with bugs package of R2OpenBUGS
Niter <- 10000
Nburn <- 1000
Nthin <- 10
################ results of descriptive statistics #############
modelo <- bugs(data = linedata,
inits = lineinits,
parameters.to.save = parameters,
model.file = linemodel,
n.chains = 1,
n.iter = Niter,
n.burnin = Nburn,
n.thin = Nthin,
debug = TRUE)
return(modelo$sims.matrix[,1] / modelo$sims.matrix[,2])
}3.2 Posterior distribution
GY <- bayes(df$GY)
GYP <- bayes(df$GYP)
HGW <- bayes(df$HGW)
TGW <- bayes(df$TGW)
HW <- bayes(df$HW)
DF <- bayes(df$DF)
PH <- bayes(df$PH)
LS <- bayes(df$LS)
NSPS <- bayes(df$NSps)
NGS <- bayes(df$NGS)3.3 Credibility intervals and mean posterior for each trait
posterior <- import("http://bit.ly/data_posterior")
conf_int_bayes <-
sapply(posterior,
function(x){
conf_int <- boa.hpd(x, 0.05)
data.frame(LCI = conf_int[[1]],
MEAN = mean(x),
UCI = conf_int[[2]])
}) %>%
t()3.4 Marginal posterior density
posterior_long <- posterior %>% pivot_longer(everything())
ggplot(posterior_long, aes(value))+
geom_density(fill = "red", alpha = 0.5, size = 0.1) +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
facet_wrap(~ name, scales = "free_y", ncol = 5) +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
labs(x = "Coefficient of variation (%)",
y = "Density")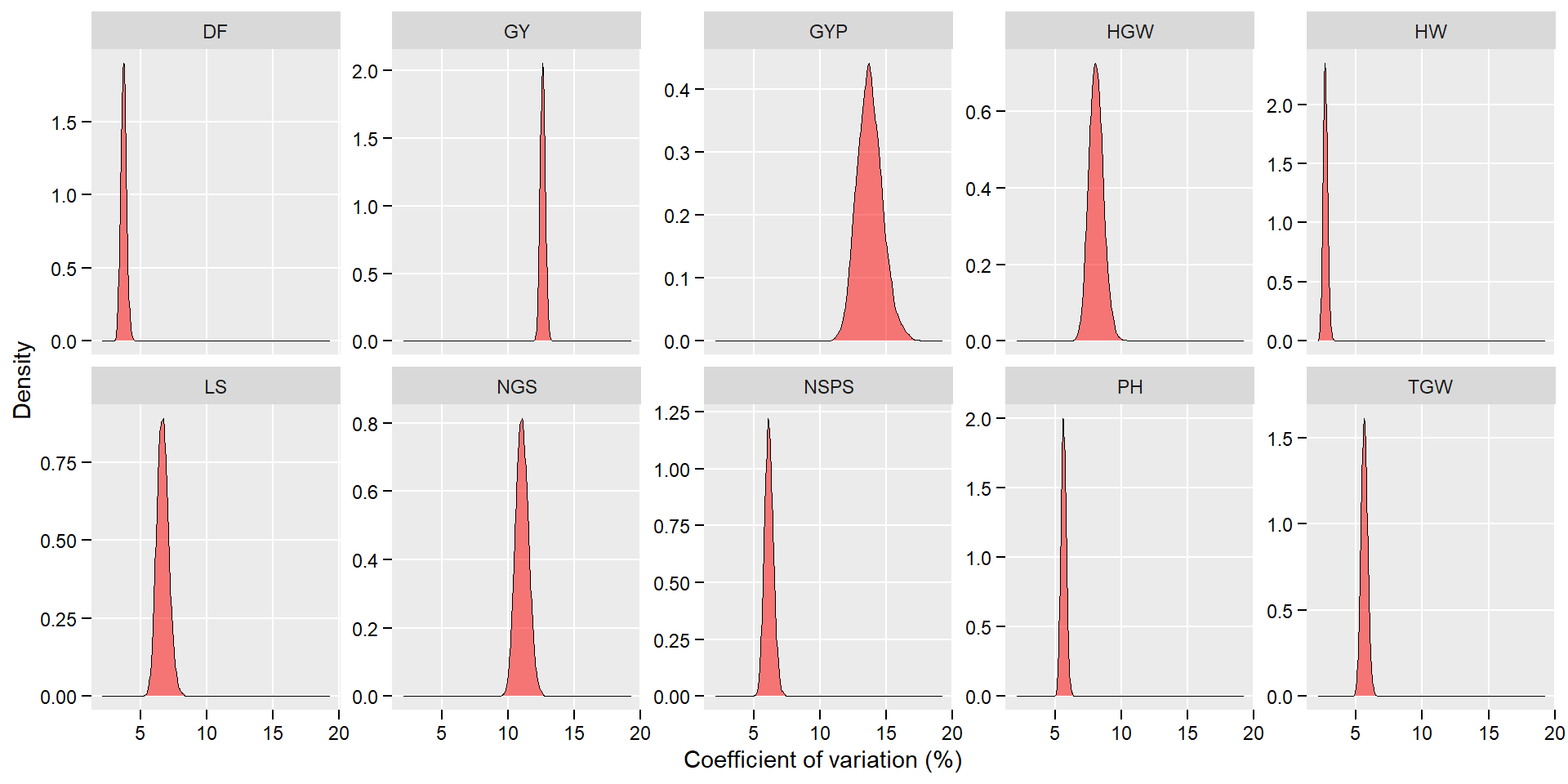
ggsave("figs/fig1_posterior.jpg", dpi = 600, width = 25, height = 10, units = "cm")
# An alternative plot
ggplot(posterior_long, aes(value))+
geom_density(aes(fill = name),
alpha = 0.5) +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
labs(x = "Coefficient of variation (%)",
y = "Density")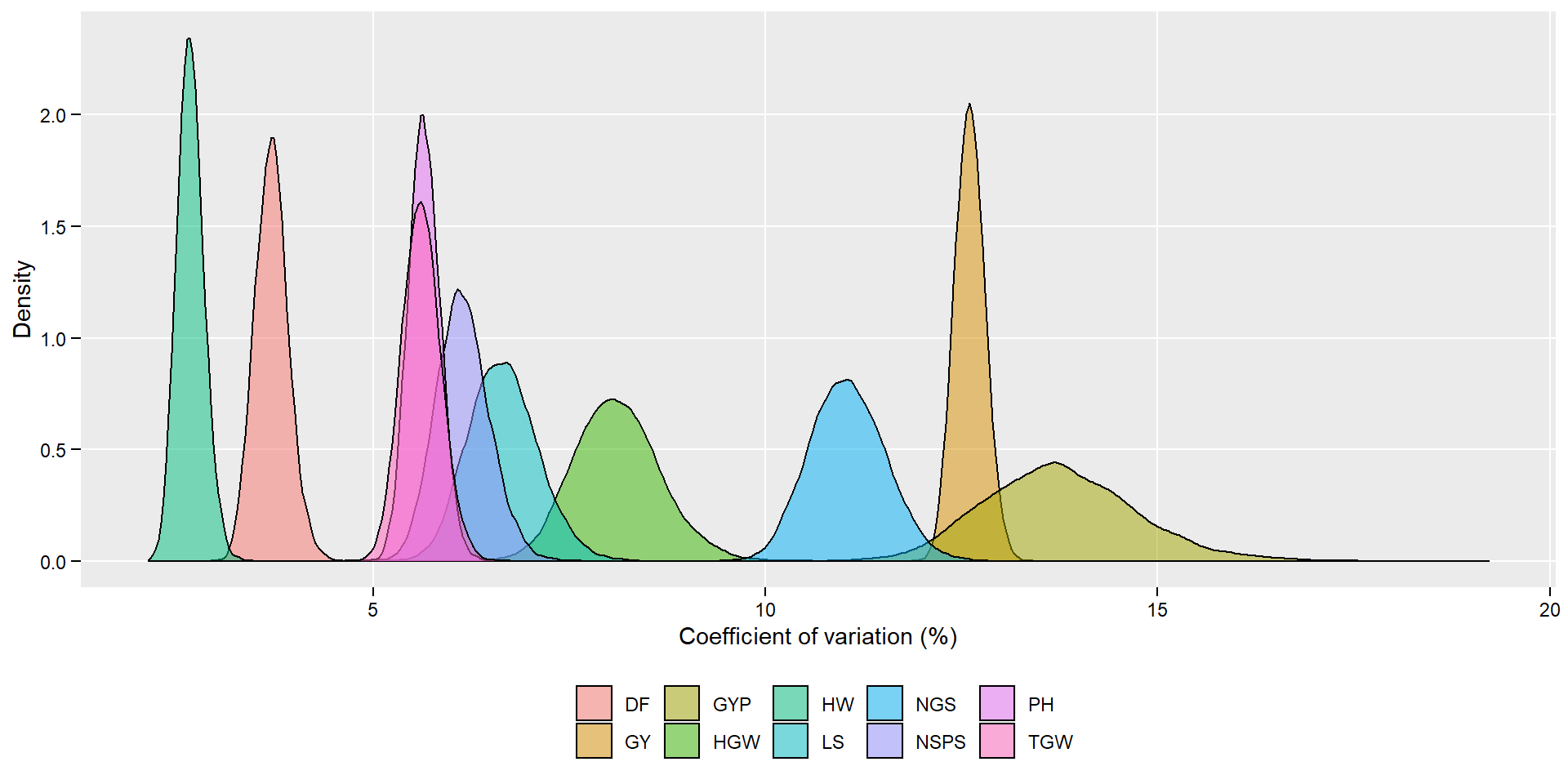
ggsave("figs/fig1_posterior2.jpg", dpi = 600, width = 25, height = 10, units = "cm")4 Frequentist
4.1 Confidence interval
get_confint <- function(df, var){
if(is.grouped_df(df)){
results <- doo(df, get_confint, var = {{var}})
return(results)
}
values <- na.omit(df %>% select_cols({{var}}) %>% pull())
model <- glm(values ~ 1, family = Gamma(link = "identity"))
conf <- confint(model)
MEAN <- coef(model)[[1]]
LCI <- conf[[1]]
UCI <- conf[[2]]
data.frame(LCI = LCI, MEAN = MEAN, UCI = UCI)
}
freq_lim <-
data_cv_long %>%
group_by(var) %>%
get_confint(cv)p <-
gf_density( ~ cv | var,
data = data_cv_long,
fill = "red",
alpha = 0.5) %>%
gf_fitdistr(linetype = 2) %>%
gf_fitdistr(dist = "gamma", color = "blue")
p +
facet_wrap(~var, nrow = 2, scales = "free_y") +
theme(panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
scale_y_continuous(expand = expansion(c(0, 0.05))) +
labs(x = "Coeficient of variation (%)",
y = "Density")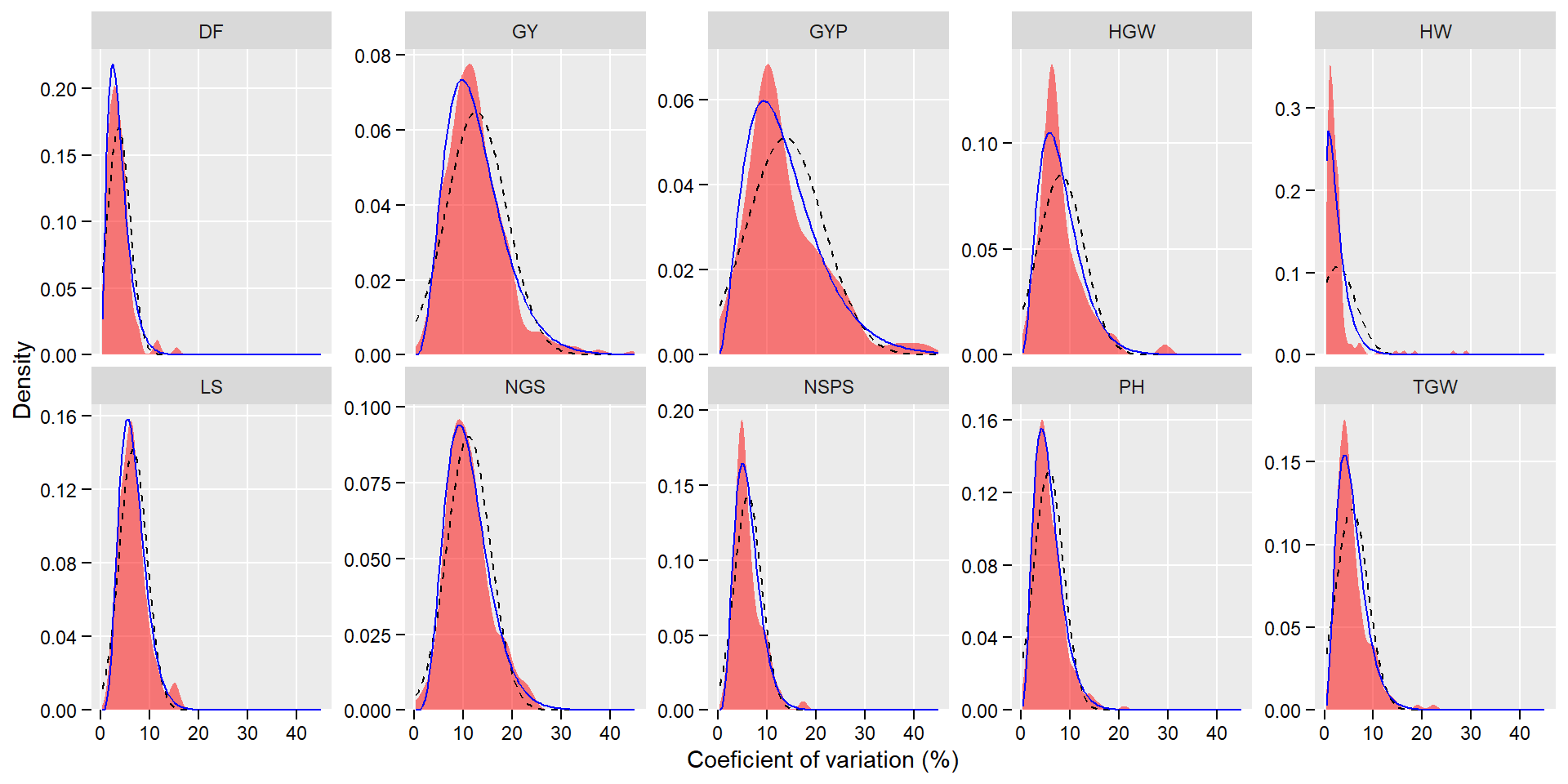
ggsave("figs/fig2_density.jpg", dpi = 600, width = 25, height = 10, units = "cm")
5 Results
5.1 Confidence interval
df_confint <- import("http://bit.ly/data_confint")
ggplot(df_confint, aes(MEAN, fct_rev(VAR), color = APPROACH)) +
geom_point(position = position_dodge(width = 0.7),
size = 2) +
geom_errorbarh(aes(xmin = LCI, xmax = UCI),
position = position_dodge(width = 0.7),
width = 0.3) +
scale_x_continuous(breaks = seq(2, 19, by = 2),
expand = c(0.15, 0.15)) +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_text(color = "black"),
axis.title = element_text(color = "black"),
axis.ticks = element_line(color = "black"),
axis.ticks.length = unit(0.15, "cm")) +
labs(x = "Coefficient of variation (%)",
y = "Variable") +
geom_text(aes(label = round(LCI, 2),
x = LCI),
position = position_dodge(width = 0.7),
hjust = 1.2,
size = 2.5,
show.legend = FALSE) +
geom_text(aes(label = round(UCI, 2),
x = UCI),
position = position_dodge(width = 0.7),
hjust = -0.3,
size = 2.5,
show.legend = FALSE)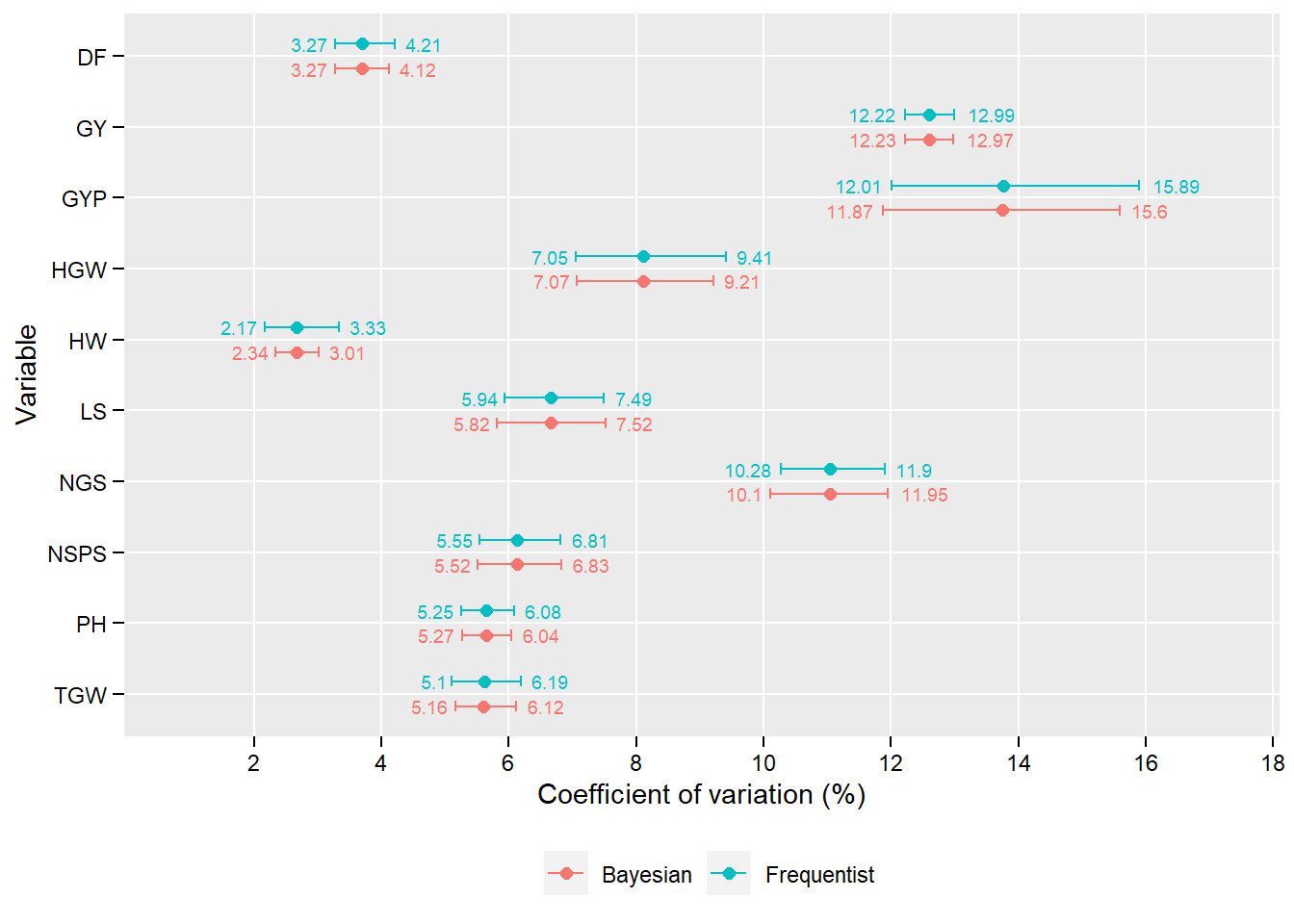
ggsave("figs/fig3_confidence.jpg", dpi = 600, width = 10, height = 12, units = "cm")JSBBbmFseXNpcyAKCmBgYHtyIGdsb2JhbF9vcHRpb25zLCBpbmNsdWRlID0gRkFMU0V9CmtuaXRyOjpvcHRzX2NodW5rJHNldChjYWNoZSA9IEZBTFNFLAogICAgICAgICAgICAgICAgICAgICAgY29tbWVudCA9ICIjIiwKICAgICAgICAgICAgICAgICAgICAgIGNvbGxhcHNlID0gVFJVRSwKICAgICAgICAgICAgICAgICAgICAgIG1lc3NhZ2UgPSBGQUxTRSwKICAgICAgICAgICAgICAgICAgICAgIHdhcm5pbmcgPSBGQUxTRSkKCmBgYAoKCiMgUGFja2FnZXMKYGBge3IgcGFja2FnZXMsIHdhcm5pbmc9RkFMU0V9CmxpYnJhcnkodGlkeXZlcnNlKQpsaWJyYXJ5KG1ldGFuKQpsaWJyYXJ5KHJpbykKbGlicmFyeShnZ3JlcGVsKQpsaWJyYXJ5KFIyT3BlbkJVR1MpCmxpYnJhcnkoYm9hKQpsaWJyYXJ5KGdnZm9ybXVsYSkKYGBgCgpUbyBwZXJmb3JtIHRoZSBiYXllc2lhbiBhbmFseXNpcywgdGhlIFtPcGVuIEJVR1NdKGh0dHBzOi8vd3d3Lm9wZW5idWdzLm5ldC93L0Rvd25sb2Fkcykgc29mdHdhcmUgbmVlZCB0byBiZSBpbnN0YWxsZWQuIEZvbGxvdyB0aGVzZSBpbnN0cnVjdGlvbnMgdG8gaW5zdGFsbCBpdC4KCgojIERhdGEKYGBge3J9CmRhdGFfY3YgPC0gIAogIGltcG9ydCgiaHR0cDovL2JpdC5seS9kYXRhX2N2cyIpICU+JSAKICBzZWxlY3QoR1k6TkdTKQoKIyBMb25nIGZvcm1hdApkYXRhX2N2X2xvbmcgPC0KZGF0YV9jdiAlPiUKICBwaXZvdF9sb25nZXIoZXZlcnl0aGluZygpLCAKICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAidmFyIiwKICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gImN2IikgJT4lCiAgcmVtb3ZlX3Jvd3NfbmEoKQoKIyBzYW1wbGVzIHBlciB2YXJpYWJsZQpkYXRhX2N2X2xvbmcgJT4lIG5fYnkodmFyKQoKIyBjcmVhdGUgYSBsaXN0IG9mIHZhcmlhYmxlcyB3aXRoIG5vIG1pc3NpbmcgdmFsdWVzCmRmIDwtIGxhcHBseShkYXRhX2N2LCByZW1vdmVfcm93c19uYSkKCmBgYAoKCgoKIyBCYXllc2lhbgojIyBGdW5jdGlvbiAKCmBgYHtyIGV2YWw9RkFMU0V9CmJheWVzIDwtIGZ1bmN0aW9uKGRmKXsKbGluZW1vZGVsIDwtIGZ1bmN0aW9uKCl7CiAgZm9yIChpIGluIDE6NjQpICMgY2hhbmdlIHRoZSBudW1iZXIgb2Ygc2FtcGxlcyBmb3IgZWFjaCB2YXJpYWJsZQogIHsKICAgIHlbaV0gfiBkZ2FtbWEociwgbXUpCiAgfSAgCiAgciB+IGR1bmlmKDAsNSkKICBtdSB+IGR1bmlmKDAsNSkgICAgCn0KIyMjIyMjIyMjIyMjIyMjIyBFc3BlY2lmaWNhbmRvIG9zIGRhZG9zCmxpbmVkYXRhIDwtIGxpc3QoeSA9IGRmW1sxXV0pCiMjIyMjIyMjIyMjIyMjIyMjIyMjIyMgU3BlY2lmaWNhdGlvbiBpbm5pdGlhbCB2YWx1ZXMKbGluZWluaXRzIDwtIGZ1bmN0aW9uKCl7bGlzdChyID0gMC41LCBtdSA9IDEpIH0KI1NwZWNpZmljYXRpb24gdGhlIHBhcmFtZXRlcnMKcGFyYW1ldGVycyA8LSBjKCJyIiwibXUiKQojIyMjIyMjIyMjIyMjIEV4ZWN1dGlvbiBmdW5jdGlvbiBhbmFseXNpcyB3aXRoIGJ1Z3MgcGFja2FnZSBvZiBSMk9wZW5CVUdTCk5pdGVyIDwtIDEwMDAwCk5idXJuIDwtIDEwMDAKTnRoaW4gPC0gMTAKIyMjIyMjIyMjIyMjIyMjIyByZXN1bHRzIG9mIGRlc2NyaXB0aXZlIHN0YXRpc3RpY3MgIyMjIyMjIyMjIyMjIwptb2RlbG8gPC0gYnVncyhkYXRhID0gbGluZWRhdGEsCiAgICAgICAgICAgICAgIGluaXRzID0gbGluZWluaXRzLAogICAgICAgICAgICAgICBwYXJhbWV0ZXJzLnRvLnNhdmUgPSBwYXJhbWV0ZXJzLAogICAgICAgICAgICAgICBtb2RlbC5maWxlID0gbGluZW1vZGVsLAogICAgICAgICAgICAgICBuLmNoYWlucyA9IDEsCiAgICAgICAgICAgICAgIG4uaXRlciA9IE5pdGVyLAogICAgICAgICAgICAgICBuLmJ1cm5pbiA9IE5idXJuLAogICAgICAgICAgICAgICBuLnRoaW4gPSBOdGhpbiwKICAgICAgICAgICAgICAgZGVidWcgPSBUUlVFKQpyZXR1cm4obW9kZWxvJHNpbXMubWF0cml4WywxXSAvIG1vZGVsbyRzaW1zLm1hdHJpeFssMl0pCn0KYGBgCgoKIyMgUG9zdGVyaW9yIGRpc3RyaWJ1dGlvbgpgYGB7ciBldmFsID0gRkFMU0V9CkdZIDwtIGJheWVzKGRmJEdZKQpHWVAgPC0gYmF5ZXMoZGYkR1lQKQpIR1cgPC0gYmF5ZXMoZGYkSEdXKQpUR1cgPC0gYmF5ZXMoZGYkVEdXKQpIVyA8LSBiYXllcyhkZiRIVykKREYgPC0gYmF5ZXMoZGYkREYpClBIIDwtIGJheWVzKGRmJFBIKQpMUyA8LSBiYXllcyhkZiRMUykKTlNQUyA8LSBiYXllcyhkZiROU3BzKQpOR1MgPC0gYmF5ZXMoZGYkTkdTKQpgYGAKCgoKIyMgQ3JlZGliaWxpdHkgaW50ZXJ2YWxzIGFuZCBtZWFuIHBvc3RlcmlvciBmb3IgZWFjaCB0cmFpdApgYGB7cn0KcG9zdGVyaW9yIDwtIGltcG9ydCgiaHR0cDovL2JpdC5seS9kYXRhX3Bvc3RlcmlvciIpCmNvbmZfaW50X2JheWVzIDwtIApzYXBwbHkocG9zdGVyaW9yLCAKICAgICAgIGZ1bmN0aW9uKHgpewogICAgICAgICBjb25mX2ludCA8LSBib2EuaHBkKHgsIDAuMDUpCiAgICAgICAgIGRhdGEuZnJhbWUoTENJID0gY29uZl9pbnRbWzFdXSwKICAgICAgICAgICAgICAgICAgICBNRUFOID0gbWVhbih4KSwKICAgICAgICAgICAgICAgICAgICBVQ0kgPSBjb25mX2ludFtbMl1dKQogICAgICAgfSkgJT4lIAogIHQoKQpgYGAKCgojIyAgTWFyZ2luYWwgcG9zdGVyaW9yIGRlbnNpdHkKYGBge3IgZmlnLndpZHRoPTEwfQpwb3N0ZXJpb3JfbG9uZyA8LSBwb3N0ZXJpb3IgJT4lIHBpdm90X2xvbmdlcihldmVyeXRoaW5nKCkpCgoKZ2dwbG90KHBvc3Rlcmlvcl9sb25nLCBhZXModmFsdWUpKSsKICBnZW9tX2RlbnNpdHkoZmlsbCA9ICJyZWQiLCBhbHBoYSA9IDAuNSwgc2l6ZSA9IDAuMSkgKwogIHRoZW1lKHBhbmVsLmdyaWQubWlub3IgPSBlbGVtZW50X2JsYW5rKCksCiAgICAgICAgbGVnZW5kLnBvc2l0aW9uID0gImJvdHRvbSIsCiAgICAgICAgbGVnZW5kLnRpdGxlID0gZWxlbWVudF9ibGFuaygpLAogICAgICAgIGF4aXMudGV4dCA9IGVsZW1lbnRfdGV4dChjb2xvciA9ICJibGFjayIpLAogICAgICAgIGF4aXMudGlja3MgPSBlbGVtZW50X2xpbmUoY29sb3IgPSAiYmxhY2siKSwKICAgICAgICBheGlzLnRpY2tzLmxlbmd0aCA9IHVuaXQoMC4xNSwgImNtIikpICsKICBmYWNldF93cmFwKH4gbmFtZSwgc2NhbGVzID0gImZyZWVfeSIsIG5jb2wgPSA1KSArCiAgdGhlbWUocGFuZWwuZ3JpZC5taW5vciA9IGVsZW1lbnRfYmxhbmsoKSwKICAgICAgICBsZWdlbmQucG9zaXRpb24gPSAiYm90dG9tIiwKICAgICAgICBsZWdlbmQudGl0bGUgPSBlbGVtZW50X2JsYW5rKCksCiAgICAgICAgYXhpcy50ZXh0ID0gZWxlbWVudF90ZXh0KGNvbG9yID0gImJsYWNrIiksCiAgICAgICAgYXhpcy50aWNrcyA9IGVsZW1lbnRfbGluZShjb2xvciA9ICJibGFjayIpLAogICAgICAgIGF4aXMudGlja3MubGVuZ3RoID0gdW5pdCgwLjE1LCAiY20iKSkgKwogIGxhYnMoeCA9ICJDb2VmZmljaWVudCBvZiB2YXJpYXRpb24gKCUpIiwKICAgICAgIHkgPSAiRGVuc2l0eSIpCgpnZ3NhdmUoImZpZ3MvZmlnMV9wb3N0ZXJpb3IuanBnIiwgZHBpID0gNjAwLCB3aWR0aCA9IDI1LCBoZWlnaHQgPSAxMCwgdW5pdHMgPSAiY20iKQoKCiMgQW4gYWx0ZXJuYXRpdmUgcGxvdApnZ3Bsb3QocG9zdGVyaW9yX2xvbmcsIGFlcyh2YWx1ZSkpKwogIGdlb21fZGVuc2l0eShhZXMoZmlsbCA9IG5hbWUpLAogICAgICAgICAgICAgICBhbHBoYSA9IDAuNSkgKwogIHRoZW1lKHBhbmVsLmdyaWQubWlub3IgPSBlbGVtZW50X2JsYW5rKCksCiAgICAgICAgbGVnZW5kLnBvc2l0aW9uID0gImJvdHRvbSIsCiAgICAgICAgbGVnZW5kLnRpdGxlID0gZWxlbWVudF9ibGFuaygpLAogICAgICAgIGF4aXMudGV4dCA9IGVsZW1lbnRfdGV4dChjb2xvciA9ICJibGFjayIpLAogICAgICAgIGF4aXMudGlja3MgPSBlbGVtZW50X2xpbmUoY29sb3IgPSAiYmxhY2siKSwKICAgICAgICBheGlzLnRpY2tzLmxlbmd0aCA9IHVuaXQoMC4xNSwgImNtIikpICsKICB0aGVtZShwYW5lbC5ncmlkLm1pbm9yID0gZWxlbWVudF9ibGFuaygpLAogICAgICAgIGxlZ2VuZC5wb3NpdGlvbiA9ICJib3R0b20iLAogICAgICAgIGxlZ2VuZC50aXRsZSA9IGVsZW1lbnRfYmxhbmsoKSwKICAgICAgICBheGlzLnRleHQgPSBlbGVtZW50X3RleHQoY29sb3IgPSAiYmxhY2siKSwKICAgICAgICBheGlzLnRpY2tzID0gZWxlbWVudF9saW5lKGNvbG9yID0gImJsYWNrIiksCiAgICAgICAgYXhpcy50aWNrcy5sZW5ndGggPSB1bml0KDAuMTUsICJjbSIpKSArCiAgbGFicyh4ID0gIkNvZWZmaWNpZW50IG9mIHZhcmlhdGlvbiAoJSkiLAogICAgICAgeSA9ICJEZW5zaXR5IikKZ2dzYXZlKCJmaWdzL2ZpZzFfcG9zdGVyaW9yMi5qcGciLCBkcGkgPSA2MDAsIHdpZHRoID0gMjUsIGhlaWdodCA9IDEwLCB1bml0cyA9ICJjbSIpCmBgYAoKCgojIEZyZXF1ZW50aXN0CiMjIENvbmZpZGVuY2UgaW50ZXJ2YWwKYGBge3Igd2FybmluZz1GQUxTRSwgbWVzc2FnZT1GQUxTRX0KCmdldF9jb25maW50IDwtIGZ1bmN0aW9uKGRmLCB2YXIpewogICAgaWYoaXMuZ3JvdXBlZF9kZihkZikpewogICAgICByZXN1bHRzIDwtIGRvbyhkZiwgZ2V0X2NvbmZpbnQsIHZhciA9IHt7dmFyfX0pCiAgICAgIHJldHVybihyZXN1bHRzKQogICAgfQogICAgdmFsdWVzIDwtIG5hLm9taXQoZGYgJT4lIHNlbGVjdF9jb2xzKHt7dmFyfX0pICU+JSBwdWxsKCkpCiAgICBtb2RlbCA8LSBnbG0odmFsdWVzIH4gMSwgZmFtaWx5ID0gR2FtbWEobGluayA9ICJpZGVudGl0eSIpKQogICAgY29uZiA8LSBjb25maW50KG1vZGVsKQogICAgTUVBTiA8LSBjb2VmKG1vZGVsKVtbMV1dCiAgICBMQ0kgPC0gY29uZltbMV1dCiAgICBVQ0kgPC0gIGNvbmZbWzJdXQogICAgZGF0YS5mcmFtZShMQ0kgPSBMQ0ksIE1FQU4gPSBNRUFOLCBVQ0kgPSBVQ0kpCiAgfQoKZnJlcV9saW0gPC0gCiAgZGF0YV9jdl9sb25nICU+JQogICAgZ3JvdXBfYnkodmFyKSAlPiUgCiAgICBnZXRfY29uZmludChjdikKCmBgYAoKCmBgYHtyIGZpZy53aWR0aD0xMH0KcCA8LQogIGdmX2RlbnNpdHkoIH4gY3YgfCB2YXIsCiAgICAgICAgICAgICAgZGF0YSA9IGRhdGFfY3ZfbG9uZywKICAgICAgICAgICAgICBmaWxsID0gInJlZCIsCiAgICAgICAgICAgICAgYWxwaGEgPSAwLjUpICU+JQogIGdmX2ZpdGRpc3RyKGxpbmV0eXBlID0gMikgJT4lCiAgZ2ZfZml0ZGlzdHIoZGlzdCA9ICJnYW1tYSIsIGNvbG9yID0gImJsdWUiKQoKcCArCiAgZmFjZXRfd3JhcCh+dmFyLCBucm93ID0gMiwgc2NhbGVzID0gImZyZWVfeSIpICsKICB0aGVtZShwYW5lbC5ncmlkLm1pbm9yID0gZWxlbWVudF9ibGFuaygpLAogICAgICAgIGF4aXMudGV4dCA9IGVsZW1lbnRfdGV4dChjb2xvciA9ICJibGFjayIpLAogICAgICAgIGF4aXMudGlja3MgPSBlbGVtZW50X2xpbmUoY29sb3IgPSAiYmxhY2siKSwKICAgICAgICBheGlzLnRpY2tzLmxlbmd0aCA9IHVuaXQoMC4xNSwgImNtIikpICsKICBzY2FsZV95X2NvbnRpbnVvdXMoZXhwYW5kID0gZXhwYW5zaW9uKGMoMCwgMC4wNSkpKSArCiAgbGFicyh4ID0gIkNvZWZpY2llbnQgb2YgdmFyaWF0aW9uICglKSIsCiAgICAgICB5ID0gIkRlbnNpdHkiKQoKZ2dzYXZlKCJmaWdzL2ZpZzJfZGVuc2l0eS5qcGciLCBkcGkgPSA2MDAsIHdpZHRoID0gMjUsIGhlaWdodCA9IDEwLCB1bml0cyA9ICJjbSIpCgoKYGBgCgoKCgojIFJlc3VsdHMKIyMgQ29uZmlkZW5jZSBpbnRlcnZhbApgYGB7cn0KZGZfY29uZmludCA8LSBpbXBvcnQoImh0dHA6Ly9iaXQubHkvZGF0YV9jb25maW50IikKCgpnZ3Bsb3QoZGZfY29uZmludCwgYWVzKE1FQU4sIGZjdF9yZXYoVkFSKSwgY29sb3IgPSBBUFBST0FDSCkpICsKICBnZW9tX3BvaW50KHBvc2l0aW9uID0gcG9zaXRpb25fZG9kZ2Uod2lkdGggPSAwLjcpLAogICAgICAgICAgICAgc2l6ZSA9IDIpICsKICBnZW9tX2Vycm9yYmFyaChhZXMoeG1pbiA9IExDSSwgeG1heCA9IFVDSSksCiAgICAgICAgICAgICAgICAgcG9zaXRpb24gPSBwb3NpdGlvbl9kb2RnZSh3aWR0aCA9IDAuNyksCiAgICAgICAgICAgICAgICAgd2lkdGggPSAwLjMpICsKICBzY2FsZV94X2NvbnRpbnVvdXMoYnJlYWtzID0gc2VxKDIsIDE5LCBieSA9IDIpLAogICAgICAgICAgICAgICAgICAgICBleHBhbmQgPSBjKDAuMTUsIDAuMTUpKSArCiAgdGhlbWUocGFuZWwuZ3JpZC5taW5vciA9IGVsZW1lbnRfYmxhbmsoKSwKICAgICAgICBsZWdlbmQucG9zaXRpb24gPSAiYm90dG9tIiwKICAgICAgICBsZWdlbmQudGl0bGUgPSBlbGVtZW50X2JsYW5rKCksCiAgICAgICAgYXhpcy50ZXh0ID0gZWxlbWVudF90ZXh0KGNvbG9yID0gImJsYWNrIiksCiAgICAgICAgYXhpcy50aXRsZSAgPSBlbGVtZW50X3RleHQoY29sb3IgPSAiYmxhY2siKSwKICAgICAgICBheGlzLnRpY2tzID0gZWxlbWVudF9saW5lKGNvbG9yID0gImJsYWNrIiksCiAgICAgICAgYXhpcy50aWNrcy5sZW5ndGggPSB1bml0KDAuMTUsICJjbSIpKSArCiAgbGFicyh4ID0gIkNvZWZmaWNpZW50IG9mIHZhcmlhdGlvbiAoJSkiLAogICAgICAgeSA9ICJWYXJpYWJsZSIpICsKICBnZW9tX3RleHQoYWVzKGxhYmVsID0gcm91bmQoTENJLCAyKSwKICAgICAgICAgICAgICAgIHggPSBMQ0kpLAogICAgICAgICAgICBwb3NpdGlvbiA9IHBvc2l0aW9uX2RvZGdlKHdpZHRoID0gMC43KSwgCiAgICAgICAgICAgIGhqdXN0ID0gMS4yLAogICAgICAgICAgICBzaXplID0gMi41LAogICAgICAgICAgICBzaG93LmxlZ2VuZCA9IEZBTFNFKSArCiAgZ2VvbV90ZXh0KGFlcyhsYWJlbCA9IHJvdW5kKFVDSSwgMiksCiAgICAgICAgICAgICAgICB4ID0gVUNJKSwKICAgICAgICAgICAgcG9zaXRpb24gPSBwb3NpdGlvbl9kb2RnZSh3aWR0aCA9IDAuNyksIAogICAgICAgICAgICBoanVzdCA9IC0wLjMsCiAgICAgICAgICAgIHNpemUgPSAyLjUsCiAgICAgICAgICAgIHNob3cubGVnZW5kID0gRkFMU0UpCgpnZ3NhdmUoImZpZ3MvZmlnM19jb25maWRlbmNlLmpwZyIsIGRwaSA9IDYwMCwgd2lkdGggPSAxMCwgaGVpZ2h0ID0gMTIsIHVuaXRzID0gImNtIikKCmBgYAoKCgo=