Analysis
1 Libraries
To reproduce the examples of this material, the R packages the following packages are needed.
library(tidyverse)
library(EnvRtype)
library(metan)
library(rio)
library(factoextra)
library(FactoMineR)
library(ggrepel)
library(sp)
library(superheat)
library(corrr)
my_theme <-
theme_bw() +
theme(panel.spacing = unit(0, "cm"),
panel.grid = element_blank(),
legend.position = "bottom")2 Datasets
2.1 Traits
me1 <- c("E3", "E4", "E5")
me2 <- c("E1", "E2", "E7")
me3 <- c("E6")
me4 <- c("E8")
df_traits <-
import("https://bit.ly/df_traits_wheat") |>
as_factor(1:6) |>
mutate(me = case_when(ENV %in% me1 ~ "ME1",
ENV %in% me2 ~ "ME2",
ENV %in% me3 ~ "ME3",
ENV %in% me4 ~ "ME4"))2.2 Climate variables
2.2.1 Scripts to gather data
df_env <- import("https://bit.ly/loc_info_wheat")
ENV <- df_env$ENV |> as_character()
LAT <- df_env$LAT
LON <- df_env$LON
ALT <- df_env$ALT
START <- df_env$START
END <- df_env$END
# see more at https://github.com/allogamous/EnvRtype
df_climate <-
get_weather(env.id = ENV,
lat = LAT,
lon = LON,
start.day = START,
end.day = END)
# GDD: Growing Degree Day (oC/day)
# FRUE: Effect of temperature on radiation use efficiency (from 0 to 1)
# T2M_RANGE: Daily Temperature Range (oC day)
# SPV: Slope of saturation vapour pressure curve (kPa.Celsius)
# VPD: Vapour pressure deficit (kPa)
# ETP: Potential Evapotranspiration (mm.day)
# PEPT: Deficit by Precipitation (mm.day)
# n: Actual duration of sunshine (hour)
# N: Daylight hours (hour)
# RTA: Extraterrestrial radiation (MJ/m^2/day)
# SRAD: Solar radiation (MJ/m^2/day)
# T2M: Temperature at 2 Meters
# T2M_MAX: Maximum Temperature at 2 Meters
# T2M_MIN: Minimum Temperature at 2 Meters
# PRECTOT: Precipitation
# WS2M: Wind Speed at 2 Meters
# RH2M: Relative Humidity at 2 Meters
# T2MDEW: Dew/Frost Point at 2 Meters
# ALLSKY_SFC_LW_DWN: Downward Thermal Infrared (Longwave) Radiative Flux
# ALLSKY_SFC_SW_DWN: All Sky Insolation Incident on a Horizontal Surface
# ALLSKY_TOA_SW_DWN: Top-of-atmosphere Insolation
# [1] "env" "ETP" "GDD" "PETP" "RH2M" "SPV"
# [8] "T2M" "T2M_MAX" "T2M_MIN" "T2M_RANGE" "T2MDEW" "VPD"
# Compute other parameters
env_data <-
df_climate %>%
as.data.frame() %>%
param_temperature(Tbase1 = 5, # choose the base temperature here
Tbase2 = 28, # choose the base temperature here
merge = TRUE) %>%
param_atmospheric(merge = TRUE) %>%
param_radiation(merge = TRUE)2.2.2 Tidy climate data
env_data <- import("https://bit.ly/df_climate_tidy")
str(env_data)
## 'data.frame': 991 obs. of 23 variables:
## $ env : chr "E1" "E1" "E1" "E1" ...
## $ LON : num -46.1 -46.1 -46.1 -46.1 -46.1 ...
## $ LAT : num -19.4 -19.4 -19.4 -19.4 -19.4 ...
## $ YEAR : int 2018 2018 2018 2018 2018 2018 2018 2018 2018 2018 ...
## $ MM : int 5 5 5 5 5 5 5 5 5 5 ...
## $ DD : int 2 3 4 5 6 7 8 9 10 11 ...
## $ DOY : int 122 123 124 125 126 127 128 129 130 131 ...
## $ YYYYMMDD : chr "05/02/2018" "05/03/2018" "05/04/2018" "05/05/2018" ...
## $ daysFromStart: int 1 2 3 4 5 6 7 8 9 10 ...
## $ tmean : num 19.5 20 20.8 20.9 21.2 ...
## $ tmax : num 27.9 28.5 29.1 28.8 28.3 ...
## $ tmin : num 12.4 11.9 13.5 14.4 15 ...
## $ prec : num 0 0 0 0.03 0.15 0.09 0 0.02 0.02 0 ...
## $ ws : num 1.81 1.13 1.5 1.74 1.5 1.96 2.19 2.66 2.7 1.71 ...
## $ rh : num 64.3 67.7 65.8 71.6 70.3 ...
## $ tdew : num 11.5 12.9 13.2 14.8 14.9 ...
## $ gdd : num 15.1 14.9 15.8 16.2 16.5 ...
## $ frue : num 0.721 0.723 0.777 0.79 0.793 ...
## $ trange : num 15.5 16.6 15.6 14.5 13.3 ...
## $ vpd : num 1.24 1.15 1.27 1.11 1.08 ...
## $ spv : num 0.146 0.146 0.156 0.158 0.158 ...
## $ etp : num 9.34 8.71 9.12 9.09 8.05 ...
## $ rta : num 28.7 28.6 28.4 28.3 28.1 ...
id_var <- names(env_data)[10:23]3 Scripts
3.1 Deviance analysis
3.1.1 Model
mod <-
waasb(df_traits,
env = ENV,
gen = LINHAGEM,
rep = BLOCO,
resp = GY,
wresp = 65) # maior peso para performance
## Evaluating trait GY |============================================| 100% 00:00:02
## ---------------------------------------------------------------------------
## P-values for Likelihood Ratio Test of the analyzed traits
## ---------------------------------------------------------------------------
## model GY
## COMPLETE NA
## GEN 3.42e-03
## GEN:ENV 1.60e-12
## ---------------------------------------------------------------------------
## All variables with significant (p < 0.05) genotype-vs-environment interaction
waasb_env <-
mod$GY$model %>%
select_cols(type, Code, Y, WAASB) %>%
subset(type == "ENV") %>%
remove_cols(type) %>%
rename(env = Code)3.1.2 BLUPs
blupge <- gmd(mod, "blupge")
blupge |>
make_mat(GEN, ENV, GY) |>
kableExtra::kable()| E1 | E2 | E3 | E4 | E5 | E6 | E7 | E8 | |
|---|---|---|---|---|---|---|---|---|
| BRS264 | 5282.212 | 4375.401 | 4594.222 | 4309.549 | 4281.875 | 1388.500 | 4780.219 | 3097.102 |
| CD151 | 5133.199 | 3981.884 | 3926.679 | 3794.016 | 3484.885 | 1319.721 | 4620.826 | 2623.267 |
| ORS1403 | 4766.968 | 3736.375 | 4220.757 | 3889.708 | 3180.280 | 1257.991 | 4654.939 | 2338.469 |
| TBIOATON | 5245.098 | 4194.017 | 4931.574 | 4342.001 | 3907.777 | 1403.818 | 4381.267 | 3334.566 |
| TBIODUQUE | 4721.233 | 4002.825 | 4320.157 | 4029.873 | 3515.728 | 1194.683 | 4173.383 | 2472.263 |
| VI09004 | 5405.136 | 3823.713 | 4219.419 | 4673.127 | 3945.283 | 1567.313 | 4063.997 | 4068.188 |
| VI09007 | 5306.519 | 4077.755 | 4036.934 | 4251.391 | 3699.024 | 1348.749 | 3712.071 | 2862.309 |
| VI09031 | 5113.092 | 3957.394 | 4254.325 | 4028.081 | 3583.433 | 1515.600 | 3624.090 | 2929.144 |
| VI09037 | 5349.630 | 4144.215 | 3984.157 | 3818.913 | 3395.113 | 1322.178 | 4082.302 | 3068.612 |
| VI130679 | 4898.978 | 3482.268 | 4240.474 | 4341.289 | 3503.989 | 1290.447 | 4090.410 | 2681.753 |
| VI130755 | 4621.478 | 3607.496 | 4299.799 | 3979.093 | 3228.663 | 1536.910 | 4279.266 | 2956.022 |
| VI130758 | 4420.186 | 3883.127 | 4110.813 | 3996.567 | 3162.743 | 1465.386 | 4025.409 | 2915.004 |
| VI131313 | 4638.819 | 3365.552 | 4406.752 | 3928.296 | 3836.338 | 1027.820 | 4130.877 | 3607.132 |
| VI14001 | 4767.310 | 3882.658 | 4113.718 | 4005.160 | 3987.910 | 1274.379 | 4414.220 | 3260.650 |
| VI14022 | 5795.451 | 3893.188 | 3657.076 | 3906.779 | 3444.757 | 1409.069 | 4351.759 | 2821.162 |
| VI14026 | 5587.829 | 4356.687 | 4478.538 | 4073.389 | 3590.865 | 1644.671 | 4746.980 | 3500.302 |
| VI14050 | 5138.250 | 3740.847 | 4237.278 | 4146.238 | 3727.650 | 1120.703 | 3846.712 | 2737.465 |
| VI14055 | 5317.182 | 3807.208 | 4461.491 | 4550.300 | 3900.639 | 1305.823 | 4654.967 | 3782.046 |
| VI14088 | 4424.901 | 3853.661 | 4527.776 | 4318.089 | 3484.264 | 1447.740 | 4055.049 | 3289.226 |
| VI14118 | 5104.537 | 4268.634 | 4110.750 | 4387.661 | 3901.315 | 1460.524 | 4242.764 | 3212.279 |
| VI14127 | 5706.292 | 4364.241 | 4603.708 | 4747.488 | 3915.401 | 1740.892 | 4482.711 | 3393.733 |
| VI14194 | 5676.327 | 4162.057 | 4961.301 | 4398.171 | 3427.092 | 1629.539 | 4427.013 | 2851.565 |
| VI14197 | 5884.131 | 4282.643 | 4379.148 | 4424.835 | 3995.823 | 1551.573 | 4602.824 | 3402.922 |
| VI14204 | 5459.974 | 3703.244 | 4151.558 | 4351.972 | 3572.006 | 1646.681 | 4380.814 | 2555.760 |
| VI14208 | 5005.025 | 3947.994 | 4307.456 | 4315.366 | 3684.816 | 1439.680 | 4344.208 | 2242.330 |
| VI14214 | 5060.277 | 4182.438 | 4368.321 | 4813.690 | 3932.955 | 1467.075 | 4521.619 | 2750.499 |
| VI14239 | 4509.855 | 3706.111 | 3816.209 | 3738.575 | 3646.618 | 1090.055 | 3869.924 | 2648.904 |
| VI14327 | 5206.957 | 3838.772 | 4342.125 | 4467.298 | 3680.383 | 1304.019 | 4322.401 | 3075.899 |
| VI14426 | 4814.134 | 3970.362 | 3994.451 | 3881.549 | 3657.549 | 1469.320 | 4270.031 | 2844.000 |
| VI14668 | 5227.392 | 3898.227 | 4148.233 | 4154.099 | 3817.169 | 1671.475 | 4182.941 | 3525.416 |
| VI14708 | 4962.562 | 3870.104 | 4414.772 | 4199.288 | 3464.495 | 1191.188 | 4292.413 | 3077.765 |
| VI14774 | 4384.071 | 4211.409 | 4495.336 | 4339.294 | 3882.483 | 1745.971 | 4368.136 | 3800.955 |
| VI14867 | 4420.342 | 4165.921 | 4757.046 | 4198.044 | 3345.108 | 1515.569 | 4374.169 | 3551.408 |
| VI14881 | 4625.981 | 3964.294 | 4359.125 | 4332.484 | 3526.458 | 1232.242 | 3906.129 | 2712.908 |
| VI14950 | 4481.994 | 3748.665 | 3625.061 | 3651.642 | 3269.540 | 1178.599 | 3753.455 | 3360.014 |
| VI14980 | 4581.815 | 4252.448 | 4684.439 | 4616.686 | 3972.237 | 1448.636 | 4538.349 | 4093.405 |
3.1.3 BLUP-based stability
indexes <- blup_indexes(mod)
kableExtra::kable(indexes$GY)| GEN | Y | HMGV | HMGV_R | RPGV | RPGV_Y | RPGV_R | HMRPGV | HMRPGV_Y | HMRPGV_R | WAASB | WAASB_R |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRS264 | 4100.680 | 3399.344 | 9 | 1.0661581 | 3986.544 | 7 | 1.0629217 | 3974.442 | 6 | 4.411137 | 13 |
| CD151 | 3569.772 | 3069.257 | 27 | 0.9581046 | 3582.514 | 27 | 0.9530240 | 3563.516 | 27 | 6.472968 | 24 |
| ORS1403 | 3431.639 | 2937.676 | 32 | 0.9262602 | 3463.442 | 34 | 0.9172999 | 3429.938 | 34 | 8.474191 | 30 |
| TBIOATON | 4039.934 | 3391.916 | 10 | 1.0567667 | 3951.428 | 8 | 1.0551504 | 3945.384 | 8 | 3.645526 | 6 |
| TBIODUQUE | 3494.970 | 2955.484 | 31 | 0.9377248 | 3506.310 | 33 | 0.9320161 | 3484.964 | 33 | 4.967330 | 15 |
| VI09004 | 4044.224 | 3507.389 | 5 | 1.0743143 | 4017.041 | 5 | 1.0640415 | 3978.629 | 5 | 9.492961 | 33 |
| VI09007 | 3637.321 | 3140.779 | 23 | 0.9751691 | 3646.321 | 20 | 0.9716251 | 3633.069 | 20 | 6.223232 | 20 |
| VI09031 | 3589.642 | 3215.725 | 17 | 0.9772566 | 3654.126 | 19 | 0.9732532 | 3639.157 | 19 | 4.376393 | 11 |
| VI09037 | 3615.978 | 3121.602 | 25 | 0.9703666 | 3628.363 | 25 | 0.9674728 | 3617.543 | 23 | 5.990468 | 19 |
| VI130679 | 3511.346 | 3035.891 | 29 | 0.9468020 | 3540.251 | 30 | 0.9438644 | 3529.267 | 29 | 3.899486 | 7 |
| VI130755 | 3507.908 | 3187.475 | 20 | 0.9639141 | 3604.236 | 26 | 0.9600174 | 3589.666 | 26 | 4.604697 | 14 |
| VI130758 | 3420.731 | 3116.108 | 26 | 0.9449113 | 3533.182 | 31 | 0.9420777 | 3522.586 | 30 | 5.641056 | 18 |
| VI131313 | 3579.175 | 2912.168 | 34 | 0.9557968 | 3573.885 | 28 | 0.9391616 | 3511.683 | 31 | 8.741502 | 31 |
| VI14001 | 3705.031 | 3163.598 | 21 | 0.9904624 | 3703.505 | 18 | 0.9870455 | 3690.728 | 18 | 4.104274 | 9 |
| VI14022 | 3634.768 | 3145.821 | 22 | 0.9739454 | 3641.745 | 22 | 0.9670467 | 3615.950 | 24 | 10.242641 | 36 |
| VI14026 | 4079.307 | 3539.613 | 3 | 1.0772170 | 4027.895 | 4 | 1.0728824 | 4011.687 | 4 | 5.592509 | 17 |
| VI14050 | 3538.600 | 2935.837 | 33 | 0.9426835 | 3524.851 | 32 | 0.9366917 | 3502.447 | 32 | 4.384475 | 12 |
| VI14055 | 4046.443 | 3338.849 | 14 | 1.0559979 | 3948.553 | 9 | 1.0495848 | 3924.573 | 9 | 6.758444 | 25 |
| VI14088 | 3654.766 | 3243.731 | 16 | 0.9912060 | 3706.285 | 17 | 0.9875995 | 3692.800 | 17 | 7.181687 | 26 |
| VI14118 | 3866.786 | 3348.180 | 12 | 1.0289051 | 3847.249 | 13 | 1.0275836 | 3842.307 | 13 | 2.499678 | 2 |
| VI14127 | 4239.867 | 3670.072 | 1 | 1.1116878 | 4156.787 | 1 | 1.1091436 | 4147.274 | 1 | 5.031011 | 16 |
| VI14194 | 4005.844 | 3437.317 | 8 | 1.0544716 | 3942.846 | 10 | 1.0473564 | 3916.241 | 10 | 8.307873 | 29 |
| VI14197 | 4168.978 | 3534.048 | 4 | 1.0872193 | 4065.295 | 2 | 1.0857271 | 4059.715 | 2 | 6.309207 | 22 |
| VI14204 | 3724.131 | 3291.346 | 15 | 1.0028569 | 3749.850 | 15 | 0.9933115 | 3714.158 | 16 | 6.448952 | 23 |
| VI14208 | 3636.024 | 3126.078 | 24 | 0.9744482 | 3643.625 | 21 | 0.9625810 | 3599.252 | 25 | 8.188630 | 28 |
| VI14214 | 3934.028 | 3341.695 | 13 | 1.0364393 | 3875.420 | 12 | 1.0316511 | 3857.516 | 12 | 6.231577 | 21 |
| VI14239 | 3263.829 | 2808.385 | 36 | 0.8931993 | 3339.822 | 36 | 0.8891251 | 3324.588 | 36 | 3.265569 | 5 |
| VI14327 | 3792.597 | 3197.783 | 18 | 1.0023576 | 3747.983 | 16 | 1.0008608 | 3742.386 | 15 | 2.588900 | 3 |
| VI14426 | 3572.558 | 3190.599 | 19 | 0.9722236 | 3635.307 | 24 | 0.9703008 | 3628.117 | 21 | 3.081889 | 4 |
| VI14668 | 3856.329 | 3459.327 | 7 | 1.0411919 | 3893.191 | 11 | 1.0359063 | 3873.427 | 11 | 4.297511 | 10 |
| VI14708 | 3666.601 | 3064.466 | 28 | 0.9732026 | 3638.968 | 23 | 0.9701153 | 3627.424 | 22 | 1.819113 | 1 |
| VI14774 | 3955.560 | 3570.168 | 2 | 1.0711268 | 4005.122 | 6 | 1.0595827 | 3961.957 | 7 | 9.017335 | 32 |
| VI14867 | 3807.374 | 3356.203 | 11 | 1.0252543 | 3833.598 | 14 | 1.0177334 | 3805.476 | 14 | 9.578895 | 35 |
| VI14881 | 3532.752 | 3020.742 | 30 | 0.9498625 | 3551.695 | 29 | 0.9463929 | 3538.721 | 28 | 3.968622 | 8 |
| VI14950 | 3270.862 | 2900.714 | 35 | 0.9059717 | 3387.580 | 35 | 0.9004934 | 3367.096 | 35 | 8.053883 | 27 |
| VI14980 | 4113.677 | 3493.171 | 6 | 1.0845239 | 4055.216 | 3 | 1.0746910 | 4018.450 | 3 | 9.544612 | 34 |
3.1.4 GGE
mod_gge <- gge(blupge, ENV, GEN, GY, svp = 1)
p1 <-
plot(mod_gge,
size.text.gen = 2.5,
size.text.env = 2.5) +
my_theme
p2 <-
plot(mod_gge,
type = 2,
size.text.gen = 2.5,
size.text.env = 2.5) +
my_theme
p3 <-
plot(mod_gge,
type = 3,
size.text.gen = 2.5,
size.text.env = 2.5,
size.text.win = 3.5) +
my_theme
p4 <-
plot(mod_gge,
type = 4,
size.text.gen = 2.5,
size.text.env = 2.5,
size.text.win = 3.5) +
my_theme
p5 <-
plot(mod_gge,
type = 6,
size.text.gen = 2.5,
size.text.env = 2.5,
size.text.win = 3.5) +
my_theme
p6 <-
plot(mod_gge,
type = 8,
size.text.gen = 2.5,
size.text.env = 2.5,
size.text.win = 3.5) +
my_theme
arrange_ggplot(p1, p2, p3, p4, p5, p6,
ncol = 3,
tag_levels = "a",
guides = "collect")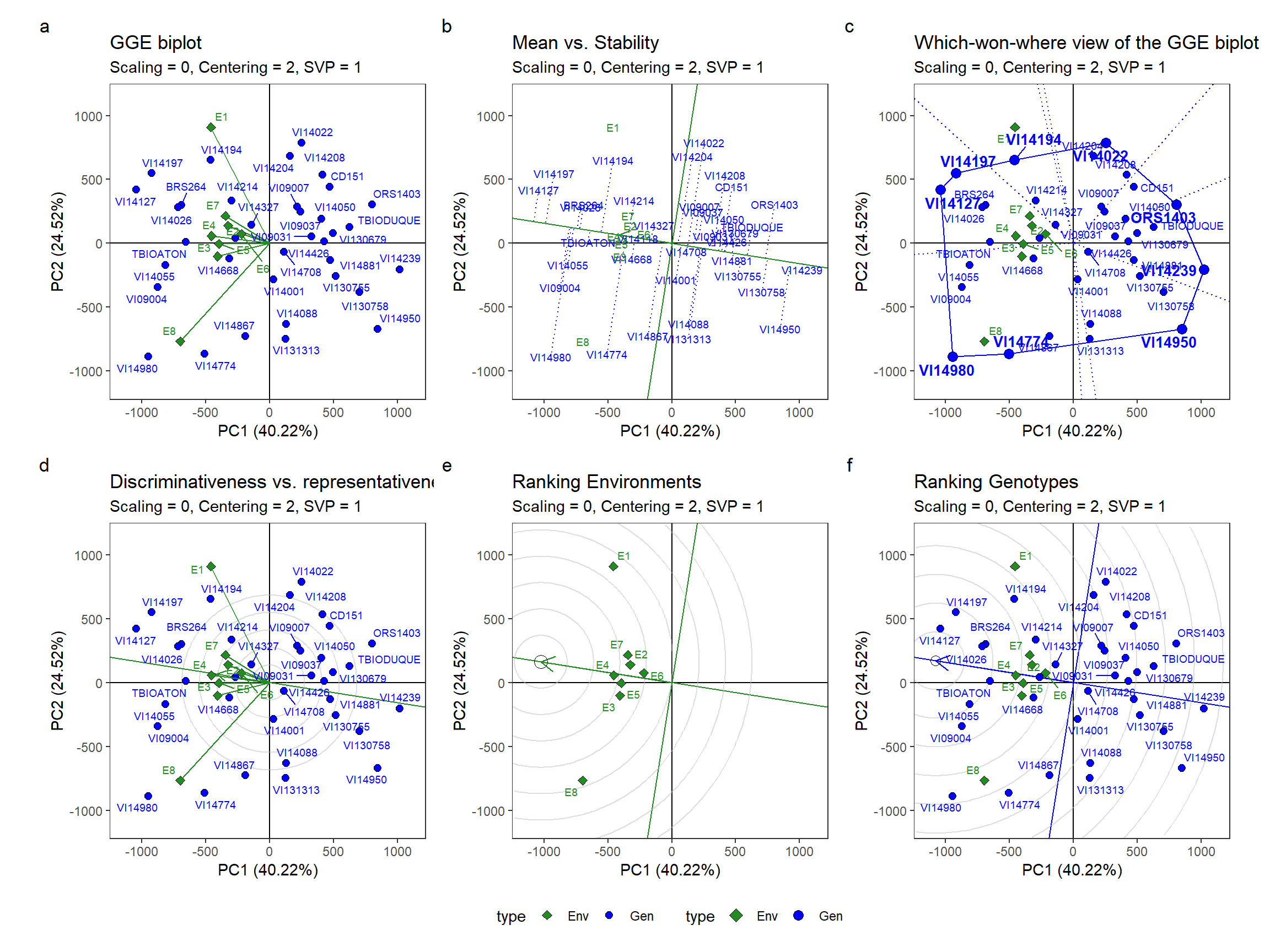
ggsave("figs/fig5_gge.png",
width = 12,
height = 9)3.1.5 WAASBY
plot_waasby(mod, size.tex.lab = 6) +
my_theme +
theme(legend.position = c(0.8, 0.1))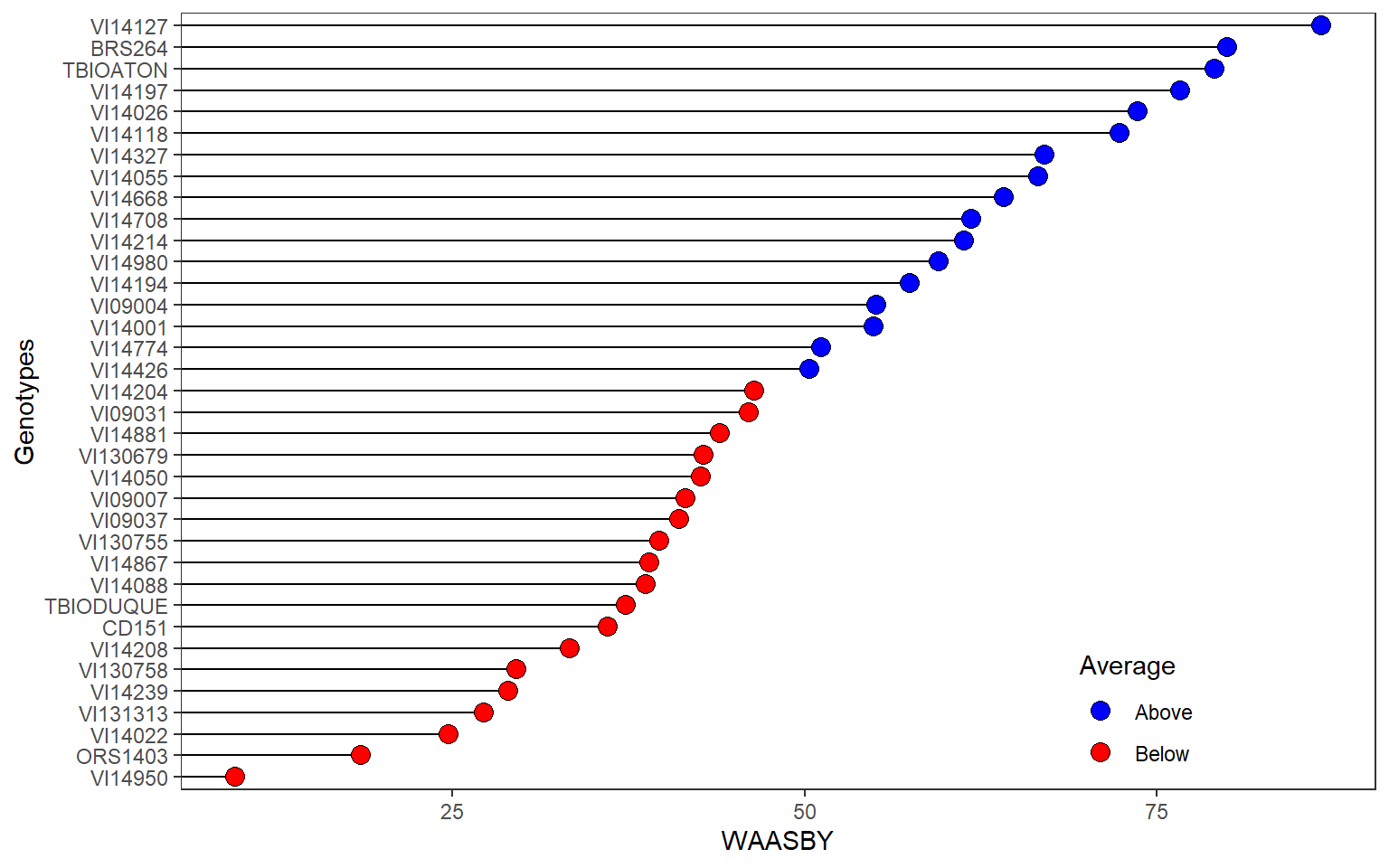
ggsave("figs/fig6_waasby.png",
width = 5,
height = 6)3.2 Correlation between climate variables
env_data |>
select_cols(tmean:rta) |>
correlate() |>
network_plot() +
guides(color = guide_colorbar(barheight = 1,
barwidth = 20,
ticks.colour = "black")) +
theme(legend.position = "bottom")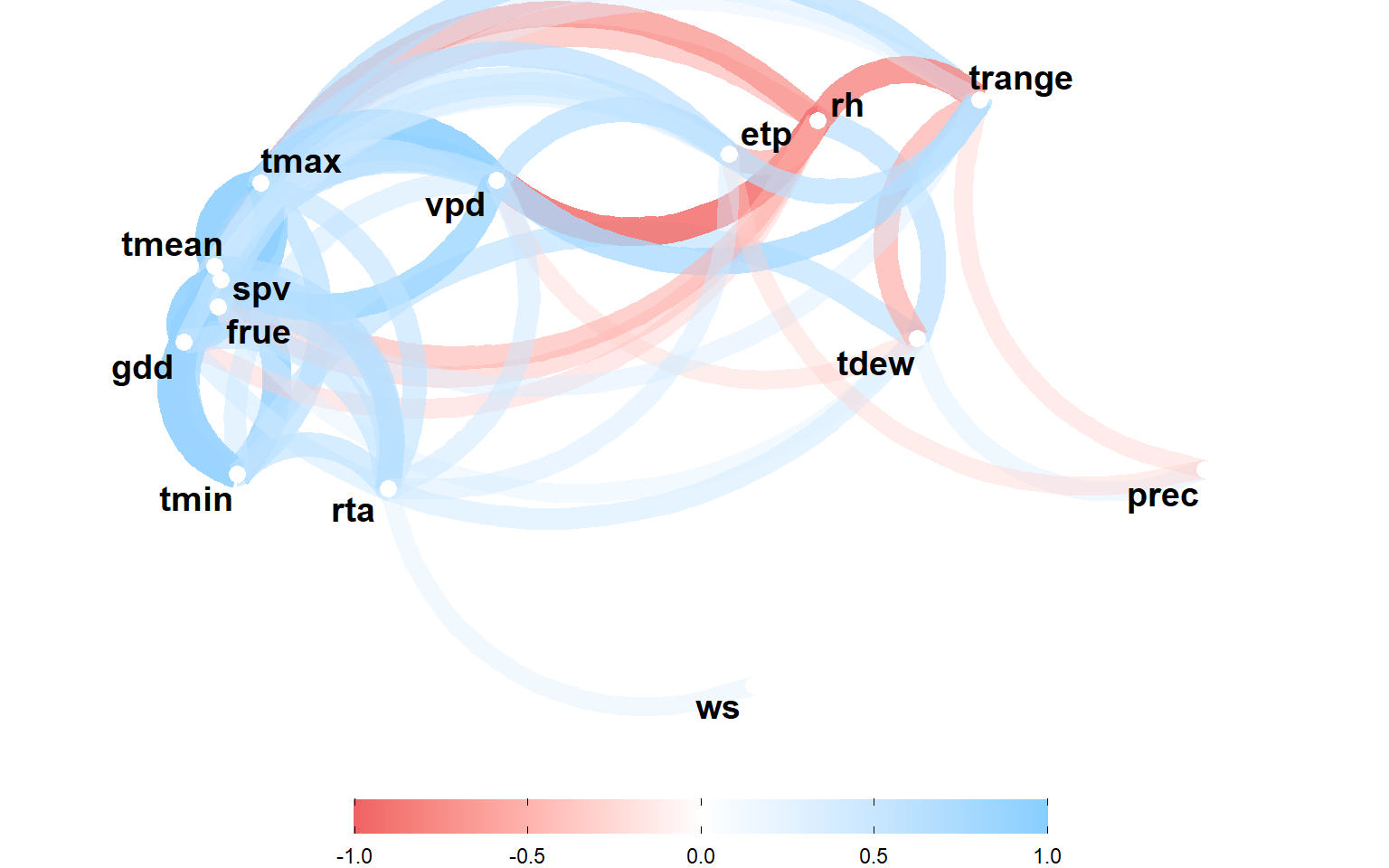
ggsave("figs/fig_network.png", width = 8, height = 8)3.3 Environmental kinships
EC <- W_matrix(env.data = env_data,
by.interval = TRUE,
statistic = 'quantile',
time.window = c(0, 30, 55, 70, 95, 130))
distances <-
env_kernel(env.data = EC,
gaussian = TRUE)
d <-
superheat(distances[[2]],
heat.pal = c("#b35806", "white", "#542788"),
pretty.order.rows = TRUE,
pretty.order.cols = TRUE,
col.dendrogram = TRUE,
legend.width = 2,
left.label.size = 0.1,
bottom.label.text.size = 5,
bottom.label.size = 0.2,
bottom.label.text.angle = 90,
legend.text.size = 17,
heat.lim = c(0, 1),
padding = 1,
legend.height=0.2)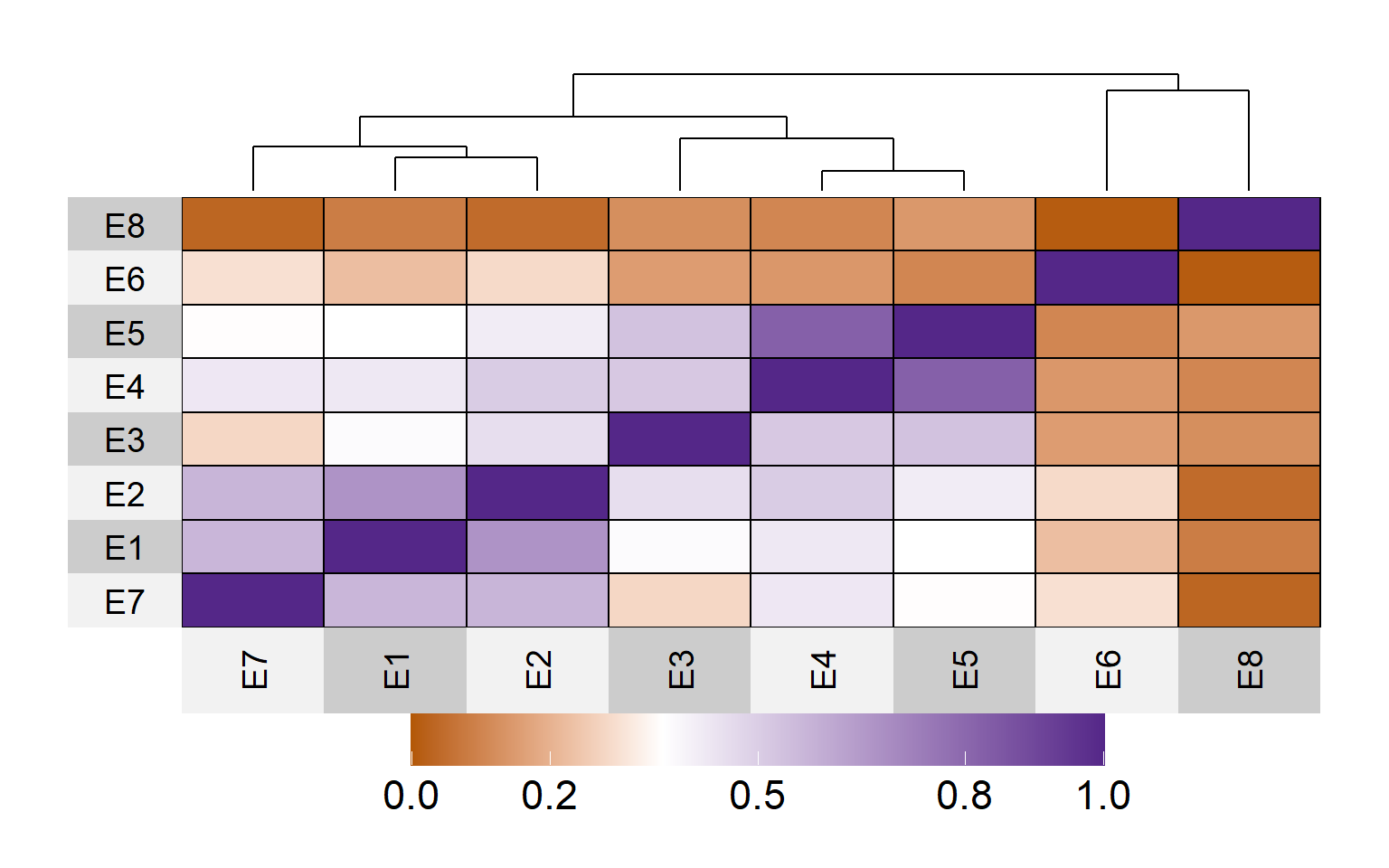
ggsave(filename = "figs/fig2_heat_env.png",
plot = d$plot,
width = 6,
height = 6)3.4 Principal Component Analysis
prec <-
env_data %>%
remove_cols(LON:YYYYMMDD, daysFromStart) |>
group_by(env) %>%
summarise(prec = sum(prec))
# compute the mean by environment and year
df_long <-
env_data %>%
remove_cols(LON:YYYYMMDD, daysFromStart) |>
remove_cols(prec) %>%
pivot_longer(-env)
# bind environment WAASB, GY, and climate traits
pca <-
df_long %>%
means_by(env, name) %>%
pivot_wider(names_from = name, values_from = value) %>%
left_join(waasb_env |> rename(GY = Y), by = "env") %>%
left_join(prec, by = "env") %>%
mutate(me = case_when(env %in% me1 ~ "ME1",
env %in% me2 ~ "ME2",
env %in% me3 ~ "ME3",
env %in% me4 ~ "ME4")) |>
column_to_rownames("env")
# compute the PCA with
pca_model <- PCA(pca,
quali.sup = 17,
graph = FALSE)
fviz_pca_biplot(pca_model,
repel = TRUE,
habillage = 17,
# font.main = c(8, "bold", "red"),
geom.var = c("arrow", "text"),
title = NULL) +
my_theme +
theme(legend.title = element_blank())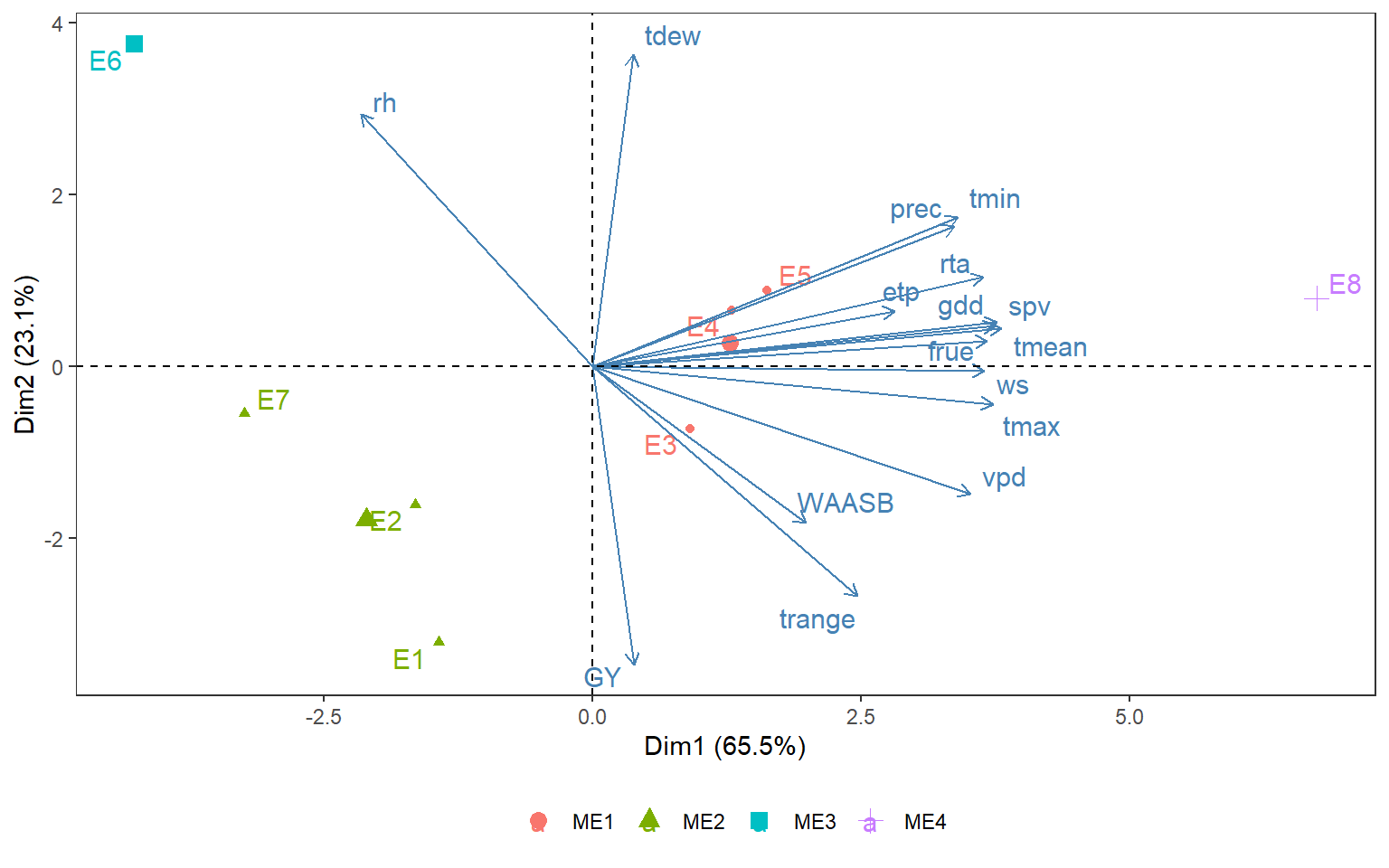
ggsave("figs/fig3_pca.png", width = 4, height = 4)3.5 Environmental tipology
names.window <-
c('1-Tillering',
'2-Boosting',
'3-heading/flowering',
'4-kernel milk stage',
'5-physiological maturity',
"")
out <-
env_typing(env.data = env_data,
env.id = "env",
var.id = c("tmax", "vpd", "rta", "etp", "rh"),
by.interval = TRUE,
time.window = c(0, 30, 55, 70, 95, 130),
names.window = names.window)
out2 <-
separate(out,
env.variable,
into = c("var", "freq"),
sep = "_",
extra = "drop") |>
mutate(me = case_when(env %in% me1 ~ "ME1",
env %in% me2 ~ "ME2",
env %in% me3 ~ "ME3",
env %in% me4 ~ "ME4"))
# plot the distribution of envirotypes
variable <- "tmax"
p1 <-
out2 |>
subset(var == variable) |> # change the variable here
ggplot() +
geom_bar(aes(x=Freq, y=env,fill=freq),
position = "fill",
stat = "identity",
width = 1,
color = "white",
size=.2)+
facet_grid(me~interval, scales = "free", space = "free")+
scale_y_discrete(expand = c(0,0))+
scale_x_continuous(expand = c(0,0))+
xlab('Relative Frequency of the maximum air temperature (ºC)')+
ylab("Environment")+
labs(fill='Envirotype')+
theme(axis.title = element_text(size=12),
legend.text = element_text(size=9),
strip.text = element_text(size=12),
legend.title = element_text(size=12),
strip.background = element_rect(fill="gray95",size=1),
legend.position = 'bottom')
# by mega environment
p2 <-
out2 |>
subset(var == variable) |> # change the variable here
sum_by(me, freq, interval) |>
ggplot() +
geom_bar(aes(x=Freq, y=me,fill=freq),
position = "fill",
stat = "identity",
width = 1,
color = "white",
size=.2)+
facet_wrap(~interval, nrow = 1)+
scale_y_discrete(expand = c(0,0))+
scale_x_continuous(expand = c(0,0))+
xlab('Relative Frequency of the maximum air temperature (ºC)')+
ylab("Mega Environment")+
labs(fill='Envirotype')+
theme(axis.title = element_text(size=12),
legend.text = element_text(size=9),
strip.text = element_text(size=12),
legend.title = element_text(size=12),
strip.background = element_rect(fill="gray95",size=1),
legend.position = 'bottom') +
scale_fill_discrete(direction = 1)
arrange_ggplot(p1, p2,
heights = c(0.6, 0.4),
tag_levels = "a",
guides = "collect")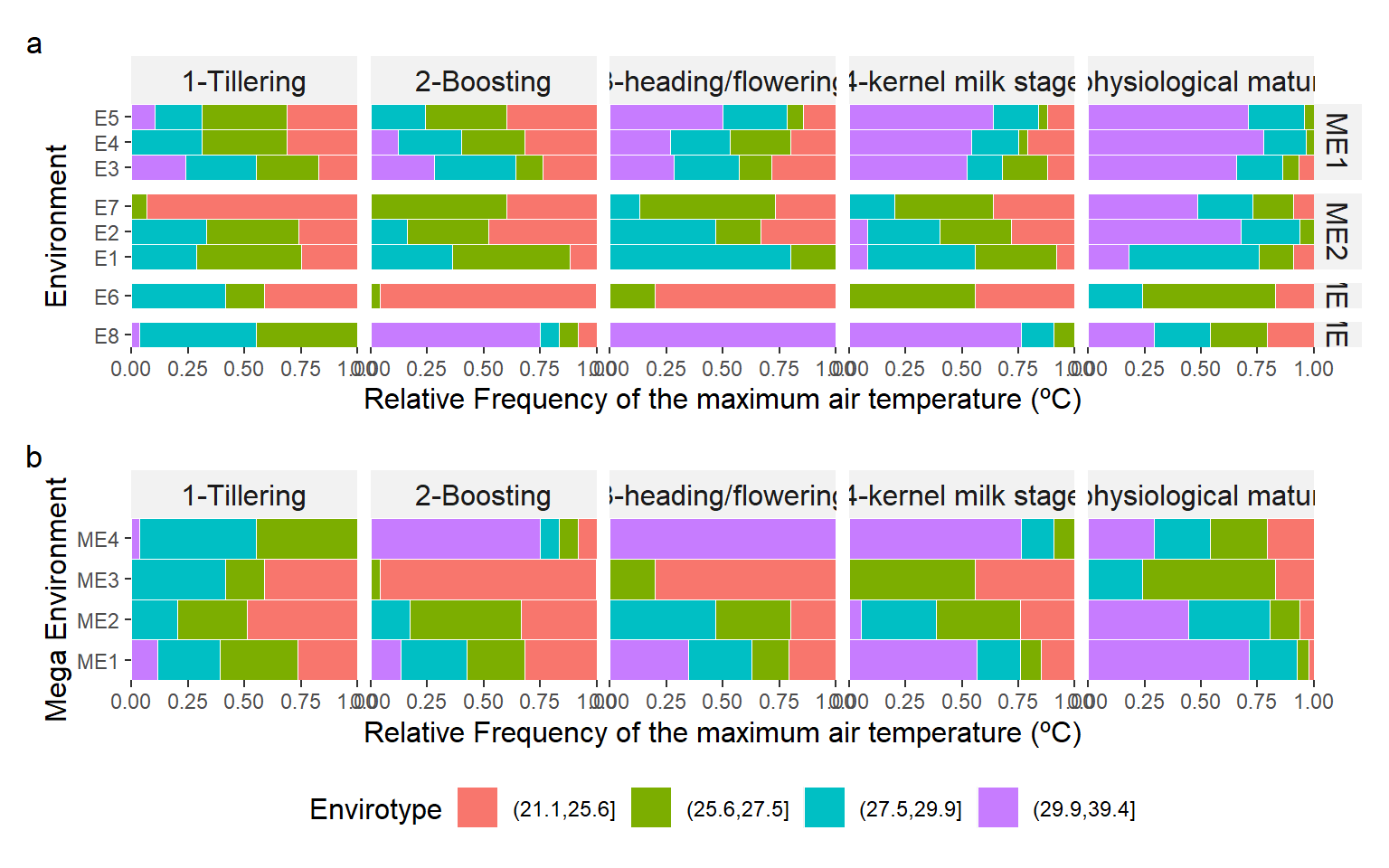
ggsave("figs/fig4_typology_tmax.png", width = 12, height = 7)4 Section info
sessionInfo()
## R version 4.2.2 (2022-10-31 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 22621)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Portuguese_Brazil.utf8 LC_CTYPE=Portuguese_Brazil.utf8
## [3] LC_MONETARY=Portuguese_Brazil.utf8 LC_NUMERIC=C
## [5] LC_TIME=Portuguese_Brazil.utf8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] corrr_0.4.4 superheat_0.1.0 sp_1.5-1 ggrepel_0.9.2
## [5] FactoMineR_2.7 factoextra_1.0.7 rio_0.5.29 metan_1.17.0.9000
## [9] EnvRtype_1.1.0 forcats_0.5.2 stringr_1.5.0 dplyr_1.0.10
## [13] purrr_0.3.5 readr_2.1.3 tidyr_1.2.1 tibble_3.1.8
## [17] ggplot2_3.4.0 tidyverse_1.3.2
##
## loaded via a namespace (and not attached):
## [1] readxl_1.4.1 backports_1.4.1 systemfonts_1.0.4
## [4] plyr_1.8.8 splines_4.2.2 TH.data_1.1-1
## [7] digest_0.6.31 foreach_1.5.2 htmltools_0.5.4
## [10] lmerTest_3.1-3 fansi_1.0.3 magrittr_2.0.3
## [13] googlesheets4_1.0.1 cluster_2.1.4 tzdb_0.3.0
## [16] openxlsx_4.2.5.1 modelr_0.1.10 sandwich_3.0-2
## [19] svglite_2.1.0 timechange_0.1.1 rmdformats_1.0.4
## [22] colorspace_2.0-3 rvest_1.0.3 textshaping_0.3.6
## [25] haven_2.5.1 xfun_0.35 crayon_1.5.2
## [28] jsonlite_1.8.4 lme4_1.1-31 survival_3.4-0
## [31] zoo_1.8-11 iterators_1.0.14 glue_1.6.2
## [34] kableExtra_1.3.4 polyclip_1.10-4 gtable_0.3.1
## [37] gargle_1.2.1 emmeans_1.8.3 webshot_0.5.4
## [40] car_3.1-1 abind_1.4-5 scales_1.2.1
## [43] mvtnorm_1.1-3 DBI_1.1.3 GGally_2.1.2
## [46] rstatix_0.7.1 Rcpp_1.0.9 viridisLite_0.4.1
## [49] xtable_1.8-4 flashClust_1.01-2 foreign_0.8-83
## [52] DT_0.26 htmlwidgets_1.6.0 httr_1.4.4
## [55] RColorBrewer_1.1-3 ellipsis_0.3.2 pkgconfig_2.0.3
## [58] reshape_0.8.9 farver_2.1.1 multcompView_0.1-8
## [61] sass_0.4.4 dbplyr_2.2.1 utf8_1.2.2
## [64] tidyselect_1.2.0 labeling_0.4.2 rlang_1.0.6
## [67] reshape2_1.4.4 munsell_0.5.0 cellranger_1.1.0
## [70] tools_4.2.2 cachem_1.0.6 cli_3.4.1
## [73] generics_0.1.3 broom_1.0.2 mathjaxr_1.6-0
## [76] evaluate_0.19 fastmap_1.1.0 ggdendro_0.1.23
## [79] yaml_2.3.6 ragg_1.2.4 knitr_1.41
## [82] fs_1.5.2 zip_2.2.2 nlme_3.1-160
## [85] leaps_3.1 xml2_1.3.3 compiler_4.2.2
## [88] rstudioapi_0.14 curl_4.3.3 ggsignif_0.6.4
## [91] reprex_2.0.2 tweenr_2.0.2 bslib_0.4.2
## [94] stringi_1.7.8 highr_0.9 lattice_0.20-45
## [97] Matrix_1.5-3 nloptr_2.0.3 vctrs_0.5.1
## [100] pillar_1.8.1 lifecycle_1.0.3 jquerylib_0.1.4
## [103] estimability_1.4.1 data.table_1.14.6 patchwork_1.1.2
## [106] R6_2.5.1 bookdown_0.31 codetools_0.2-18
## [109] boot_1.3-28 MASS_7.3-58.1 assertthat_0.2.1
## [112] withr_2.5.0 multcomp_1.4-20 hms_1.1.2
## [115] grid_4.2.2 coda_0.19-4 minqa_1.2.5
## [118] rmarkdown_2.19 carData_3.0-5 googledrive_2.0.0
## [121] ggpubr_0.5.0 ggforce_0.4.1 numDeriv_2016.8-1.1
## [124] scatterplot3d_0.3-42 lubridate_1.9.0