Computes the percentage of symptomatic leaf area using color palettes or RGB
indexes by each leaf of an image. This allows, for example, processing
replicates of the same treatment and obtaining the results for each
replication with a single image. To do that, leaf samples are first splitten
with object_split() and then, measure_disease() is applied to the list of
leaves.
Usage
measure_disease_byl(
img,
index = "B",
index_lb = "B",
index_dh = "NGRDI",
lower_size = NULL,
watershed = TRUE,
invert = FALSE,
fill_hull = FALSE,
filter = 3,
threshold = "Otsu",
extension = NULL,
tolerance = NULL,
object_size = "large",
img_healthy = NULL,
img_symptoms = NULL,
plot = TRUE,
save_image = FALSE,
dir_original = NULL,
dir_processed = NULL,
pattern = NULL,
parallel = FALSE,
workers = NULL,
show_features = FALSE,
verbose = TRUE,
...
)Arguments
- img
The image to be analyzed.
- index
A character value specifying the target mode for conversion to binary to segment the leaves from background. Defaults to "B" (blue). See
image_index()for more details. Personalized indexes can be informed as, e.g.,index = "R*G/B.- index_lb
The index used to segment the foreground (e.g., leaf) from the background. If not declared, the entire image area (pixels) will be considered in the computation of the severity.
- index_dh
The index used to segment diseased from healthy tissues when
img_healthyandimg_symptomsare not declared. Defaults to"GLI". Seeimage_index()for more details.- lower_size
To prevent dust from affecting object segmentation, objects with lesser than
10%of the mean of all objects are removed. . One can set a known area or uselower_limit = 0to select all objects (not advised).- watershed
If
TRUE(default) performs watershed-based object detection. This will detect objects even when they are touching one other. IfFALSE, all pixels for each connected set of foreground pixels are set to a unique object. This is faster but is not able to segment touching objects.- invert
Inverts the binary image if desired. This is useful to process images with a black background. Defaults to
FALSE. Ifreference = TRUEis use,invertcan be declared as a logical vector of length 2 (eg.,invert = c(FALSE, TRUE). In this case, the segmentation of objects and reference from the foreground usingback_fore_indexis performed using the default (not inverted), and the segmentation of objects from the reference is performed by inverting the selection (selecting pixels higher than the threshold).- fill_hull
Fill holes in the binary image? Defaults to
FALSE. This is useful to fill holes in objects that have portions with a color similar to the background. IMPORTANT: Objects touching each other can be combined into one single object, which may underestimate the number of objects in an image.- filter
Performs median filtering in the binary image? See more at
image_filter(). Defaults toFALSE. Use a positive integer to define the size of the median filtering. Larger values are effective at removing noise, but adversely affect edges.- threshold
The theshold method to be used.
By default (
threshold = "Otsu"), a threshold value based on Otsu's method is used to reduce the grayscale image to a binary image. If a numeric value is informed, this value will be used as a threshold.If
threshold = "adaptive", adaptive thresholding (Shafait et al. 2008) is used, and will depend on thekandwindowsizearguments.If any non-numeric value different than
"Otsu"and"adaptive"is used, an iterative section will allow you to choose the threshold based on a raster plot showing pixel intensity of the index.
- extension
Radius of the neighborhood in pixels for the detection of neighboring objects. Higher value smooths out small objects.
- tolerance
The minimum height of the object in the units of image intensity between its highest point (seed) and the point where it contacts another object (checked for every contact pixel). If the height is smaller than the tolerance, the object will be combined with one of its neighbors, which is the highest.
- object_size
The size of the object. Used to automatically set up
toleranceandextensionparameters. One of the following."small"(e.g, wheat grains),"medium"(e.g, soybean grains),"large"(e.g, peanut grains), and"elarge"(e.g, soybean pods)`.- img_healthy
A color palette of healthy tissues.
- img_symptoms
A color palette of lesioned tissues.
- plot
Show image after processing?
- save_image
Save the image after processing? The image is saved in the current working directory named as
proc_*where*is the image name given inimg.- dir_original, dir_processed
The directory containing the original and processed images. Defaults to
NULL. In this case, the function will search for the imageimgin the current working directory. After processing, whensave_image = TRUE, the processed image will be also saved in such a directory. It can be either a full path, e.g.,"C:/Desktop/imgs", or a subfolder within the current working directory, e.g.,"/imgs".- pattern
A pattern of file name used to identify images to be processed. For example, if
pattern = "im"all images that the name matches the pattern (e.g., img1.-, image1.-, im2.-) will be analyzed. Providing any number as pattern (e.g.,pattern = "1") will select images that are named as 1.-, 2.-, and so on.- parallel
Processes the images asynchronously (in parallel) in separate R sessions running in the background on the same machine. It may speed up the processing time, especially when
patternis used is informed. The number of sections is set up to 30% of available cores.- workers
A positive numeric scalar or a function specifying the maximum number of parallel processes that can be active at the same time.
- show_features
If
TRUEreturnS the lesion features such as number, area, perimeter, and radius. Defaults toFALSE.- verbose
If
TRUE(default) a summary is shown in the console.- ...
Additional arguments passed on to
measure_disease().
Value
A list with the following objects:
severityA data frame with the percentage of healthy and symptomatic areas for each leaf in the image(s).shape,statisticsIfshow_features = TRUEis used, returns the shape (area, perimeter, etc.) for each lesion and a summary statistic of the results.
Examples
library(pliman)
img <- image_pliman("mult_leaves.jpg", plot = TRUE)
 sev <-
measure_disease_byl(img = img,
index_lb = "B",
index_dh = "NGRDI",
workers = 2)
sev <-
measure_disease_byl(img = img,
index_lb = "B",
index_dh = "NGRDI",
workers = 2)
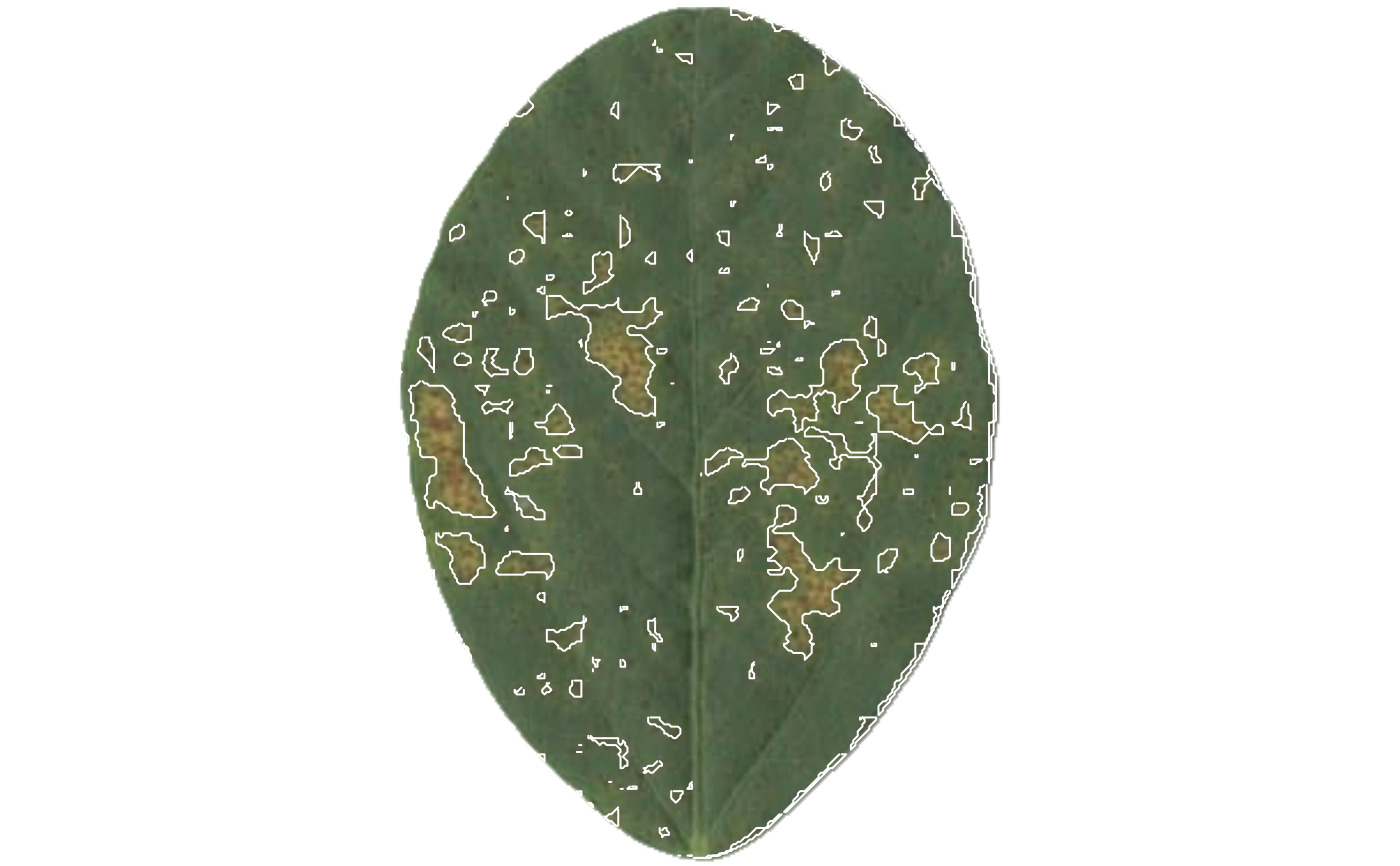

 sev$severity
#> img leaf healthy symptomatic
#> 1 img 1 84.36079 15.63921
#> 2 img 2 79.76666 20.23334
#> 3 img 3 68.39437 31.60563
sev$severity
#> img leaf healthy symptomatic
#> 1 img 1 84.36079 15.63921
#> 2 img 2 79.76666 20.23334
#> 3 img 3 68.39437 31.60563
