Compute the Mahalanobis distance of all pairwise rows in .means. The
result is a symmetric matrix containing the distances that may be used for
hierarchical clustering.
Arguments
- .means
A matrix of data with, say, p columns.
- covar
The covariance matrix.
- inverted
Logical argument. If
TRUE,covaris supposed to contain the inverse of the covariance matrix.
Author
Tiago Olivoto tiagoolivoto@gmail.com
Examples
# \donttest{
library(metan)
library(dplyr)
# Compute the mean for genotypes
means <- mean_by(data_ge, GEN) %>%
column_to_rownames("GEN")
# Compute the covariance matrix
covmat <- cov(means)
# Compute the distance
dist <- mahala(means, covmat)
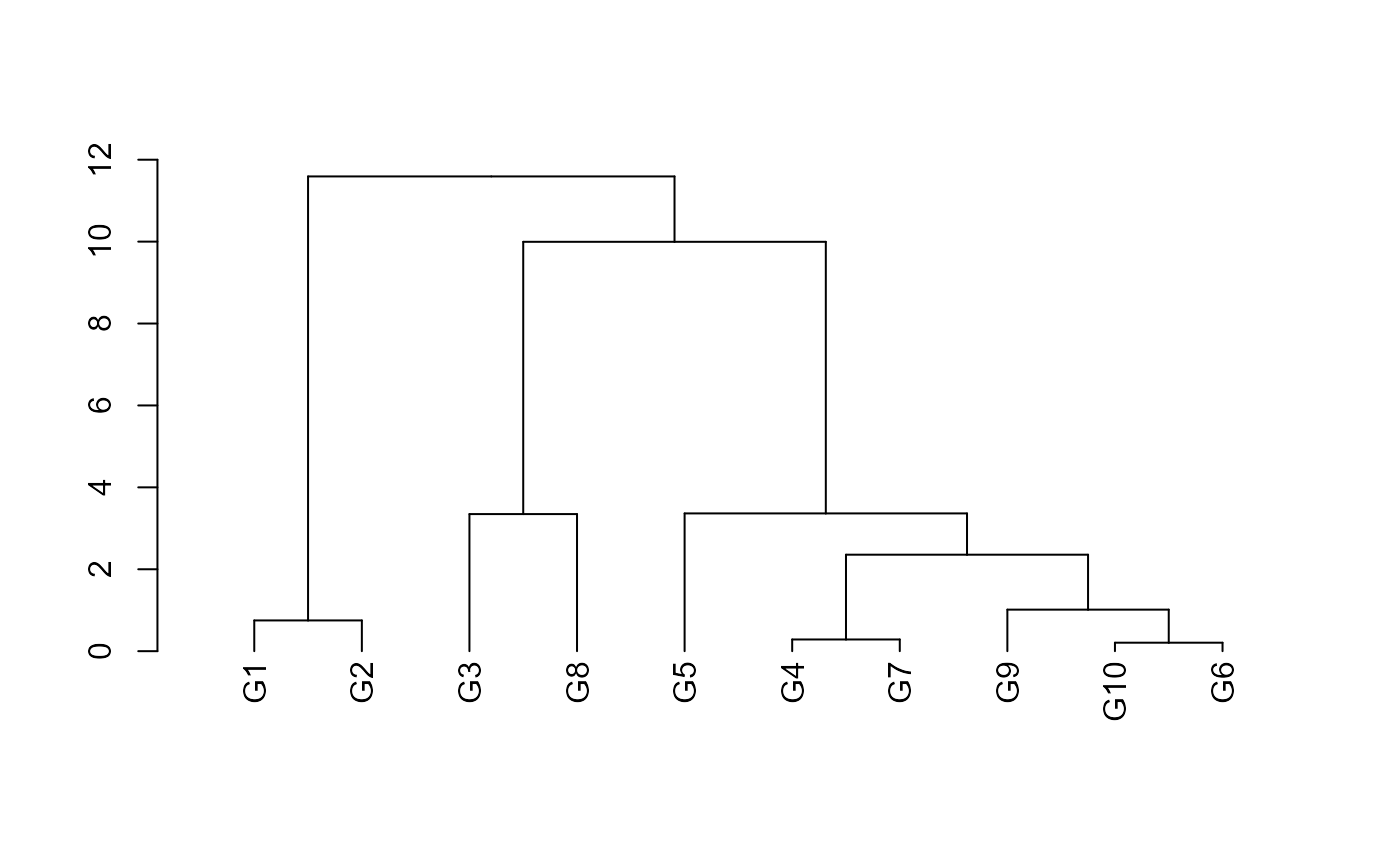
# Dendrogram
dend <- dist %>%
as.dist() %>%
hclust() %>%
as.dendrogram()
plot(dend)
 # }
# }
