01: Desempenho agronômico do linho em diferentes épocas de semeadura
1 Pacotes
To reproduce the examples of this material, the R packages the following packages are needed.
2 Dados
df_p <-
import("data/data_parcelas.csv")3 Análise de variância
3.1 Rendimento de grãos
modrg <- with(df_p,
PSUBDBC(epoca, cultivar, bloco, rgha,
xlab = "Épocas de semeadura"))
##
## -----------------------------------------------------------------
## Normality of errors
## -----------------------------------------------------------------
## Method Statistic p.value
## Shapiro-Wilk normality test(W) 0.8962594 0.01794671
##
##
##
## -----------------------------------------------------------------
## Homogeneity of Variances
## -----------------------------------------------------------------
## Plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 27.67379 4.252128e-06
##
##
## ----------------------------------------------------
## Split-plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 2.030255 0.1541947
##
##
## ----------------------------------------------------
## Interaction
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 23.33425 0.001490467
##
##
## -----------------------------------------------------------------
## Additional Information
## -----------------------------------------------------------------
##
## CV1 (%) = 14.97
## CV2 (%) = 34.61
## Mean = 743.825
## Median = 533.9
##
## -----------------------------------------------------------------
## Analysis of Variance
## -----------------------------------------------------------------
## Df Sum Sq Mean Sq F value Pr(>F)
## F1 3 6881050.432 2293683.477 185.0987045 p<0.001
## Block 2 2873.868 1436.934 0.1159596 0.892
## Error A 6 74350.066 12391.678
## F2 1 4298.727 4298.727 0.0648632 0.805
## F1 x F2 3 105689.020 35229.673 0.5315782 0.673
## Error B 8 530189.913 66273.739
## -----------------------------------------------------------------
## No significant interaction
## -----------------------------------------------------------------
##
## -----------------------------------------------------------------
## F1
## -----------------------------------------------------------------
## Multiple Comparison Test: Tukey HSD
## resp groups
## E1 1469.8333 a
## E2 1045.8667 b
## E4 236.1667 c
## E3 223.4333 c
##
##
## -----------------------------------------------------------------
## Isolated factors 2 not significant
## -----------------------------------------------------------------
## Mean
## D 730.4417
## M 757.2083
rg <-
modrg$graph1 +
labs(x = "Épocas de semeadura",
y = expression(Rendimento~de~grãos~(kg~ha^{-1})))3.2 Massa de mil grãos
modmmg <- with(df_p,
PSUBDBC(epoca, cultivar, bloco, mmg))
##
## -----------------------------------------------------------------
## Normality of errors
## -----------------------------------------------------------------
## Method Statistic p.value
## Shapiro-Wilk normality test(W) 0.9587968 0.4147291
##
##
##
## -----------------------------------------------------------------
## Homogeneity of Variances
## -----------------------------------------------------------------
## Plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 1.522785 0.6770217
##
##
## ----------------------------------------------------
## Split-plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 0.3166758 0.5736123
##
##
## ----------------------------------------------------
## Interaction
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 7.564106 0.3725931
##
##
## -----------------------------------------------------------------
## Additional Information
## -----------------------------------------------------------------
##
## CV1 (%) = 17.2
## CV2 (%) = 11.93
## Mean = 4.8387
## Median = 4.9463
##
## -----------------------------------------------------------------
## Analysis of Variance
## -----------------------------------------------------------------
## Df Sum Sq Mean Sq F value Pr(>F)
## F1 3 22.2493125 7.4164375 10.7060422 0.008
## Block 2 0.6355228 0.3177614 0.4587063 0.653
## Error A 6 4.1564029 0.6927338
## F2 1 0.9561335 0.9561335 2.8715898 0.129
## F1 x F2 3 2.3794528 0.7931509 2.3820984 0.145
## Error B 8 2.6637050 0.3329631
## -----------------------------------------------------------------
## No significant interaction
## -----------------------------------------------------------------
##
## -----------------------------------------------------------------
## F1
## -----------------------------------------------------------------
## Multiple Comparison Test: Tukey HSD
## resp groups
## E2 5.857779 a
## E1 5.743322 a
## E4 3.898390 b
## E3 3.855251 b
##
##
## -----------------------------------------------------------------
## Isolated factors 2 not significant
## -----------------------------------------------------------------
## Mean
## D 5.038282
## M 4.639089
mmg <-
modmmg$graph1 +
labs(x = "Épocas de semeadura",
y = "Massa de mil grãos (g)")
arrange_ggplot(rg, mmg,
tag_levels = "a")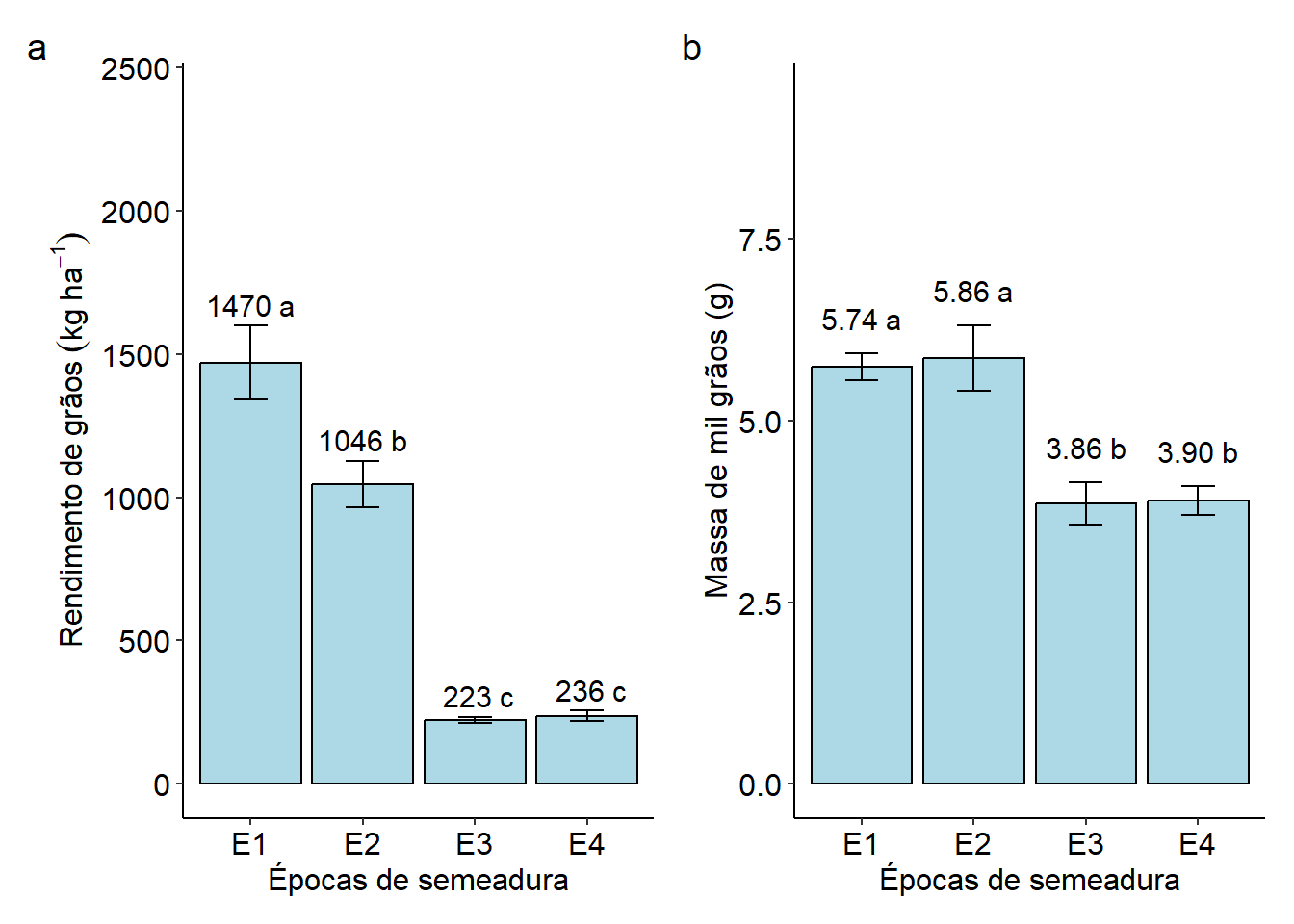
ggsave("figs/rg_mmg.jpg",
width = 8,
height = 4)3.3 Número de cápsulas
modnc <- with(df_p,
PSUBDBC(epoca, cultivar, bloco, nc))
##
## -----------------------------------------------------------------
## Normality of errors
## -----------------------------------------------------------------
## Method Statistic p.value
## Shapiro-Wilk normality test(W) 0.9498939 0.2695177
##
##
##
## -----------------------------------------------------------------
## Homogeneity of Variances
## -----------------------------------------------------------------
## Plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 1.510963 0.6797424
##
##
## ----------------------------------------------------
## Split-plot
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 0.0277831 0.8676198
##
##
## ----------------------------------------------------
## Interaction
## Method Statistic p.value
## Bartlett test(Bartlett's K-squared) 2.553598 0.9230073
##
##
## -----------------------------------------------------------------
## Additional Information
## -----------------------------------------------------------------
##
## CV1 (%) = 32.15
## CV2 (%) = 9.72
## Mean = 21.7385
## Median = 21
##
## -----------------------------------------------------------------
## Analysis of Variance
## -----------------------------------------------------------------
## Df Sum Sq Mean Sq F value Pr(>F)
## F1 3 1076.18417 358.72806 7.3462116 0.020
## Block 2 38.21202 19.10601 0.3912624 0.692
## Error A 6 292.99025 48.83171
## F2 1 66.32847 66.32847 14.8571981 0.005
## F1 x F2 3 45.72412 15.24137 3.4139807 0.073
## Error B 8 35.71520 4.46440
## -----------------------------------------------------------------
## No significant interaction
## -----------------------------------------------------------------
##
## -----------------------------------------------------------------
## F1
## -----------------------------------------------------------------
## Multiple Comparison Test: Tukey HSD
## resp groups
## E1 31.99954 a
## E2 23.40231 ab
## E3 16.09656 b
## E4 15.45556 b
##
##
##
## -----------------------------------------------------------------
## F2
## -----------------------------------------------------------------
## Multiple Comparison Test: Tukey HSD
## resp groups
## D 23.40093 a
## M 20.07606 b
pnce <-
modnc$graph1 +
labs(x = "Épocas de semeadura",
y = "Número de cápsulas")
pncc <-
modnc$graph2 +
labs(x = "Cultivares",
y = "Número de cápsulas")
arrange_ggplot(pnce, pncc, widths = c(0.75, 0.25), tag_levels = "a")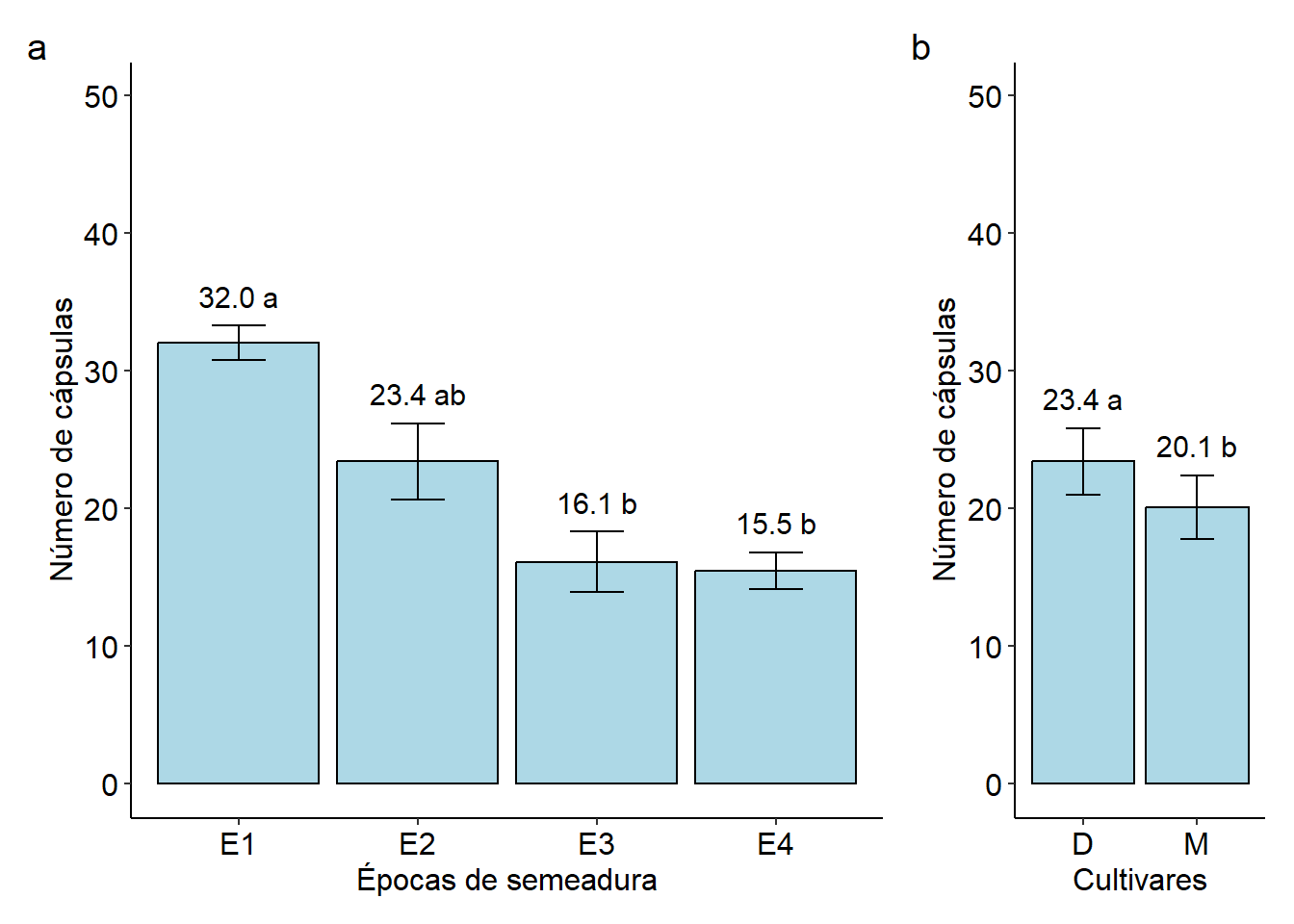
ggsave("figs/nc.jpg",
width = 8,
height = 4)4 Correlação
corr_coef(df_p, rgha, mmg, nc) |>
network_plot(legend_position = "bottom")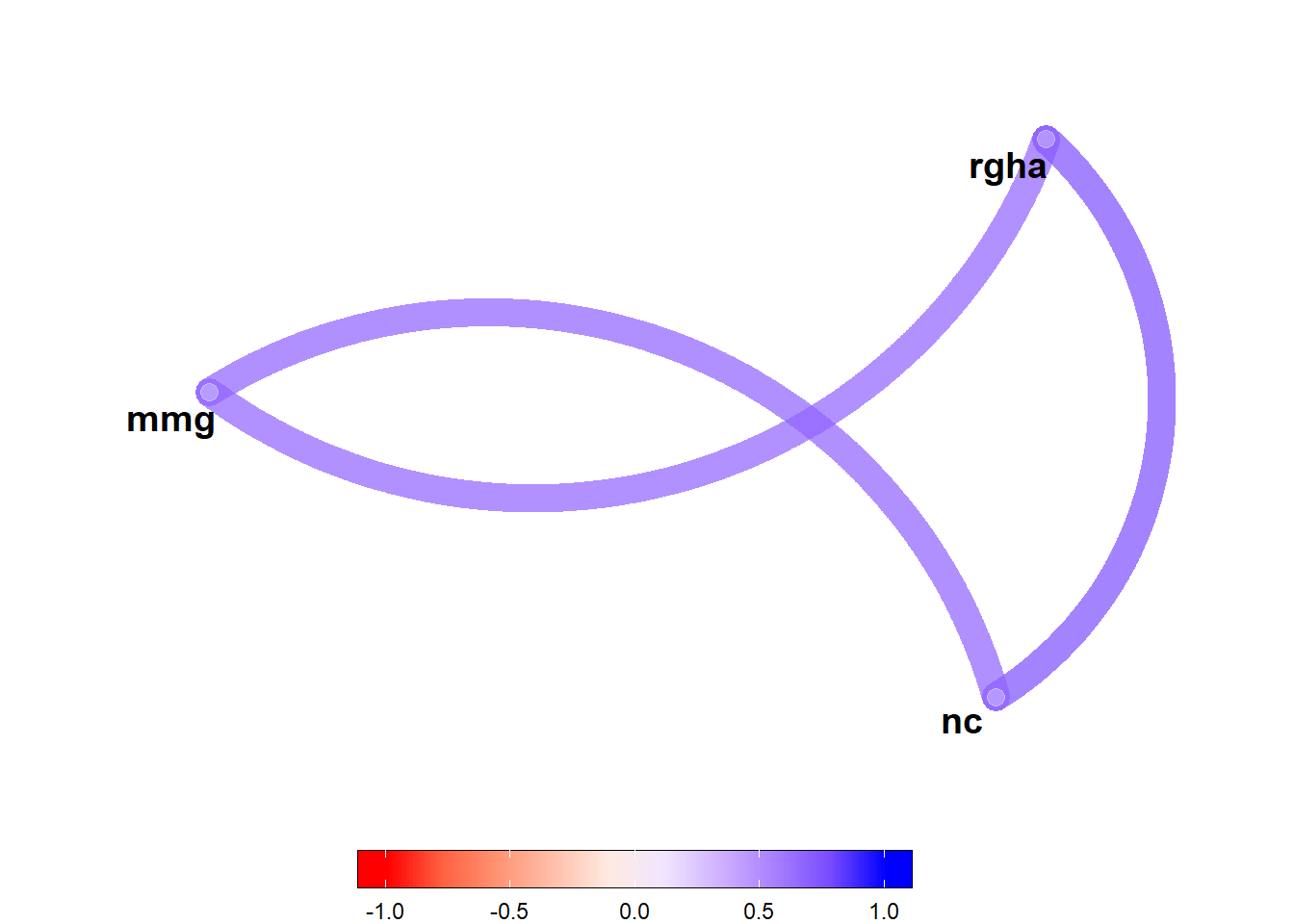
ggsave("figs/corr_epocas.jpg",
width = 10,
height = 3)5 Section info
sessionInfo()
## R version 4.2.2 (2022-10-31 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 22621)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Portuguese_Brazil.utf8 LC_CTYPE=Portuguese_Brazil.utf8
## [3] LC_MONETARY=Portuguese_Brazil.utf8 LC_NUMERIC=C
## [5] LC_TIME=Portuguese_Brazil.utf8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] metan_1.18.0 AgroR_1.3.3 lubridate_1.9.2 forcats_1.0.0
## [5] stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1 readr_2.1.4
## [9] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2 tidyverse_2.0.0
## [13] rio_0.5.29
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-160 RColorBrewer_1.1-3 numDeriv_2016.8-1.1
## [4] tools_4.2.2 utf8_1.2.3 R6_2.5.1
## [7] nortest_1.0-4 colorspace_2.1-0 withr_2.5.0
## [10] GGally_2.1.2 tidyselect_1.2.0 gridExtra_2.3
## [13] emmeans_1.8.7 curl_5.0.1 compiler_4.2.2
## [16] textshaping_0.3.6 cli_3.6.1 sandwich_3.0-2
## [19] labeling_0.4.2 scales_1.2.1 lmtest_0.9-40
## [22] mvtnorm_1.2-2 multcompView_0.1-9 systemfonts_1.0.4
## [25] digest_0.6.33 foreign_0.8-83 minqa_1.2.5
## [28] rmarkdown_2.23 pkgconfig_2.0.3 htmltools_0.5.5
## [31] lme4_1.1-34 plotrix_3.8-2 fastmap_1.1.1
## [34] dunn.test_1.3.5 htmlwidgets_1.6.2 rlang_1.1.1
## [37] readxl_1.4.3 rstudioapi_0.15.0 farver_2.1.1
## [40] generics_0.1.3 zoo_1.8-12 jsonlite_1.8.7
## [43] gtools_3.9.4 zip_2.3.0 car_3.1-2
## [46] magrittr_2.0.3 patchwork_1.1.2 Matrix_1.6-0
## [49] Rcpp_1.0.11 munsell_0.5.0 fansi_1.0.4
## [52] abind_1.4-5 lifecycle_1.0.3 stringi_1.7.12
## [55] multcomp_1.4-25 yaml_2.3.7 carData_3.0-5
## [58] mathjaxr_1.6-0 MASS_7.3-60 plyr_1.8.8
## [61] grid_4.2.2 ggrepel_0.9.3 crayon_1.5.2
## [64] lattice_0.20-45 haven_2.5.3 cowplot_1.1.1
## [67] splines_4.2.2 hms_1.1.3 knitr_1.43
## [70] pillar_1.9.0 boot_1.3-28 estimability_1.4.1
## [73] codetools_0.2-18 glue_1.6.2 drc_3.0-1
## [76] evaluate_0.21 data.table_1.14.8 tweenr_2.0.2
## [79] vctrs_0.6.3 nloptr_2.0.3 tzdb_0.4.0
## [82] cellranger_1.1.0 polyclip_1.10-4 gtable_0.3.3
## [85] reshape_0.8.9 ggforce_0.4.1 xfun_0.39
## [88] openxlsx_4.2.5.2 xtable_1.8-4 coda_0.19-4
## [91] ragg_1.2.5 survival_3.4-0 lmerTest_3.1-3
## [94] timechange_0.2.0 TH.data_1.1-2