07: Diversidade genética entre acessos de linho (Linum usitatissimum, L.) baseada em caracteres morfoagronômicos
1 Libraries
To reproduce the examples of this material, the R packages the following packages are needed.
2 Data
df <-
import("data/data_mgidi.csv") |>
remove_rows_na()3 Adjust by season effect
df_man <-
df |>
remove_cols(gen)
mod <- manova(cbind(ac, ap, nc, ng, areac, nr, mc, rgpla, icc, ngcap, mmg) ~ epoca,
data = df_man)
coefficients(mod)
## ac ap nc ng areac nr
## (Intercept) 53.989394 81.545455 33.51515 216.07576 0.39426566 1.2878788
## epocaE2 -2.636453 -5.016043 -10.20143 -76.07576 -0.01735562 -0.8368984
## epocaE3 10.352914 0.198007 -17.49592 -135.71037 -0.05770728 -0.7301865
## epocaE4 5.143333 -12.112727 -18.18788 -146.29394 -0.11073884 -0.8696970
## mc rgpla icc ngcap mmg
## (Intercept) 1.6351515 1.2174242 0.80009953 6.4669836 5.5972761
## epocaE2 -0.5247594 -0.4180125 -0.06885532 -0.2516304 0.2900347
## epocaE3 -1.1145746 -0.9076166 -0.21017662 -1.5260380 -1.7910373
## epocaE4 -1.2189697 -0.9523333 -0.16804186 -1.9838140 -1.7085045
df_resi <-
residuals(mod) |>
as.data.frame() |>
mutate(gen = df$gen) |>
column_to_rownames("gen")4 Distances
dists <- clustering(df_resi,
scale = TRUE,
clustmethod = "average",
nclust = 16)
fviz_dend(dists$hc,
k = 16,
cex = 0.5,
repel = TRUE,
type = "circular") +
theme_void()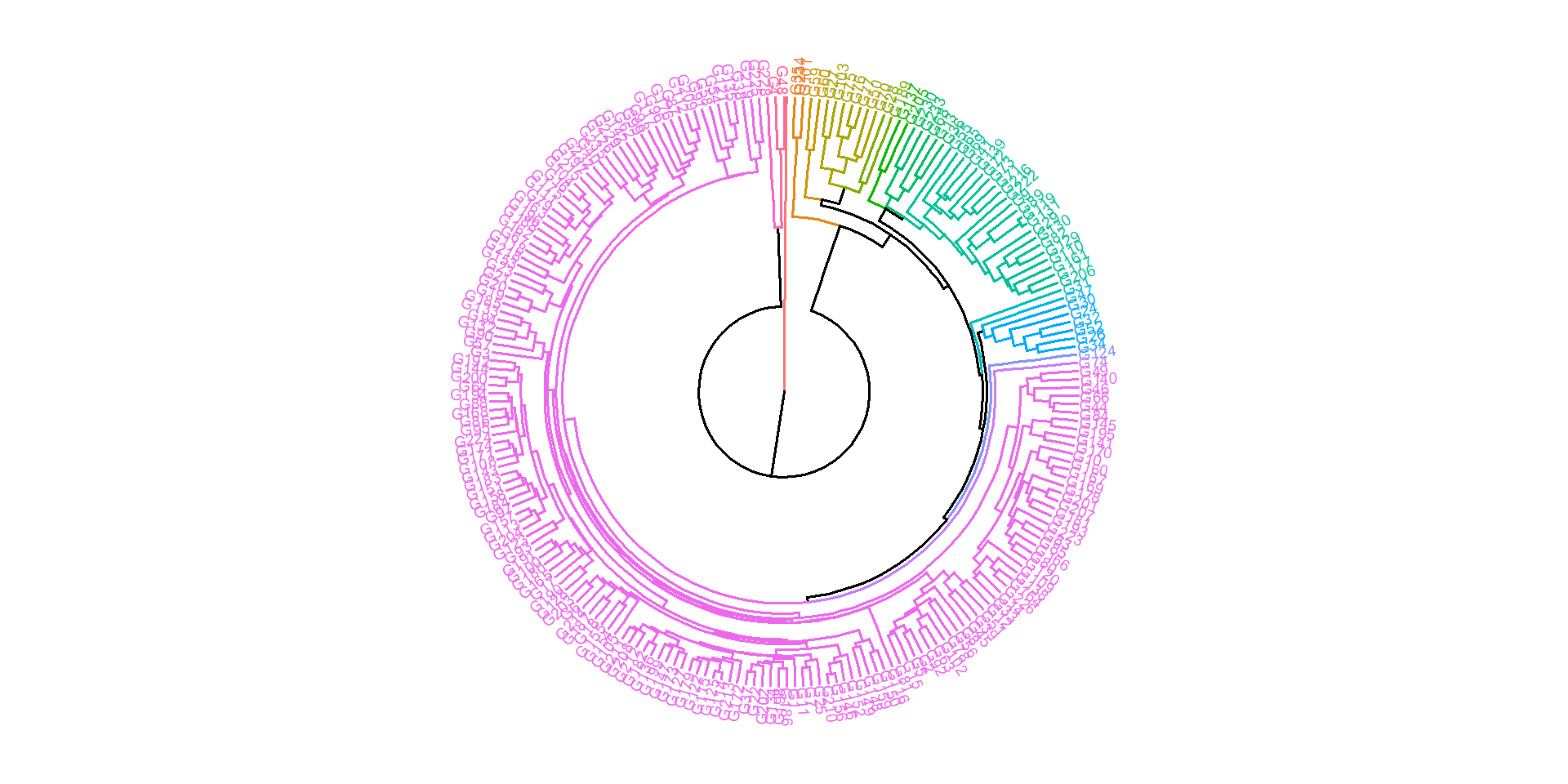
ggsave("figs/phylo.jpg",
dpi = 600,
width = 10,
height = 10)5 Médias
df2 <-
df |>
mutate(groups = dists$data |> pull(groups)) |>
as_factor(groups)
df3 <-
df2 |>
select(groups, mmg, rgpla, ng) |>
pivot_longer(-groups) |>
filter(groups %in% c(2, 3, 4, 11, 15)) |>
group_by(groups, name) |>
summarise(mean = mean(value),
sd = sd(value),
n = n(),
se = sd / sqrt(n))
df_mean <-
df |>
mean_by(.vars = c(mmg, rgpla, ng)) |>
pivot_longer(everything())
df_meansel <-
df3 |>
mean_by(name)6 Stats
ggplot(df3, aes(x = groups, y = mean, fill = groups)) +
geom_col() +
geom_hline(data = df_mean,
aes(yintercept = value),
linetype = 3,
linewidth = 1) +
geom_hline(data = df_meansel,
aes(yintercept = mean),
linetype = 8,
color = "blue",
linewidth = 1) +
geom_errorbar(aes(ymin = mean - se,
ymax = mean + se),
width = 0.2) +
facet_wrap(~name, scales = "free") +
theme_bw(base_size = 18) +
theme(legend.position = "bottom") +
scale_y_continuous(expand = expansion(c(0, 0.1))) +
geom_text(aes(y = 0, label = glue::glue("n = {n}")),
vjust = -1) +
labs(x = "Grupos",
y = "Valor médio do caractere",
fill = "Grupos")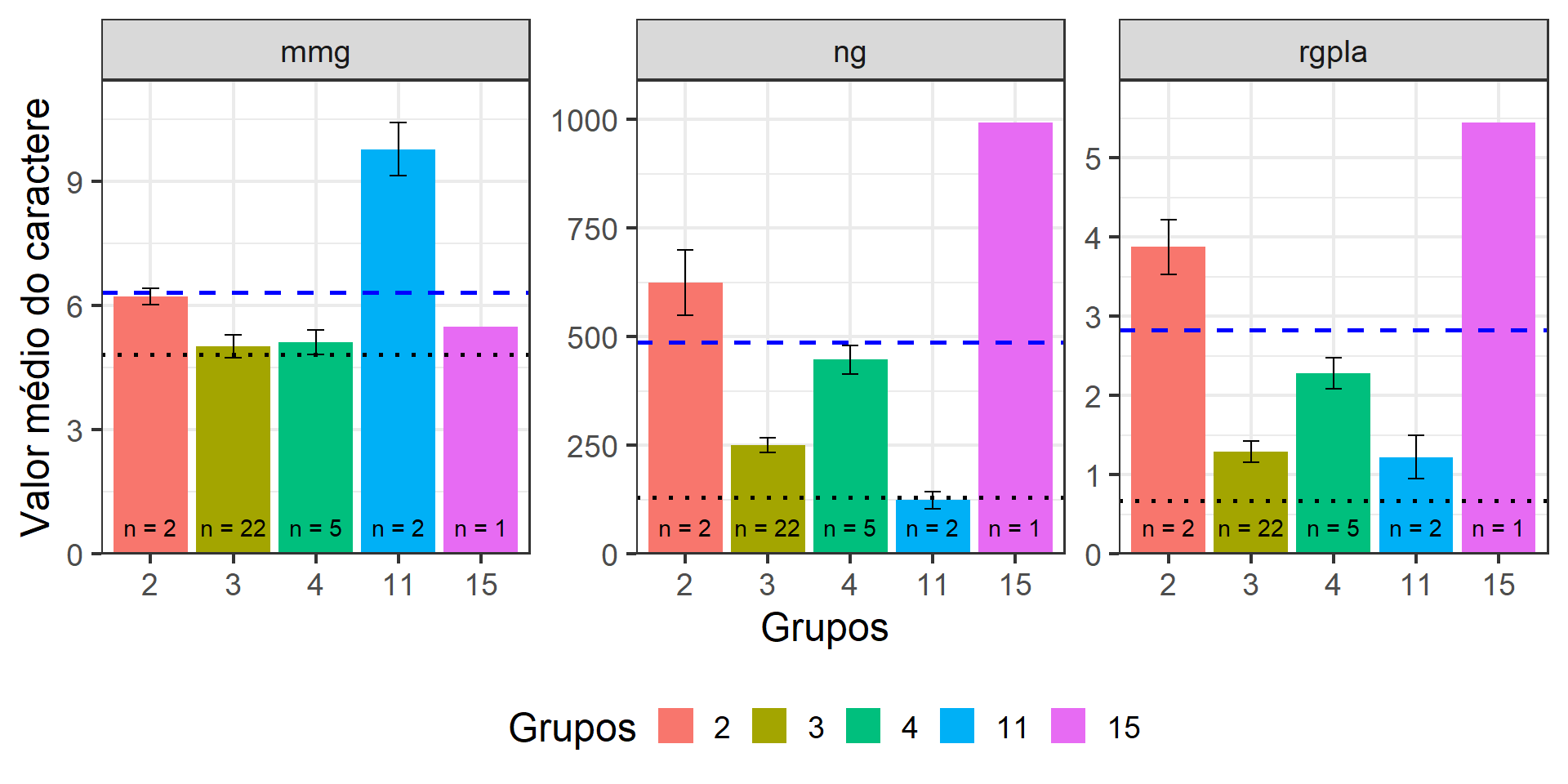
ggsave("figs/mean_clusters.jpg",
dpi = 600,
width = 12,
height = 6)7 Section info
sessionInfo()
## R version 4.2.2 (2022-10-31 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 22621)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Portuguese_Brazil.utf8 LC_CTYPE=Portuguese_Brazil.utf8
## [3] LC_MONETARY=Portuguese_Brazil.utf8 LC_NUMERIC=C
## [5] LC_TIME=Portuguese_Brazil.utf8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggstatsplot_0.11.1 factoextra_1.0.7 metan_1.18.0 lubridate_1.9.2
## [5] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1
## [9] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2
## [13] tidyverse_2.0.0 rio_0.5.29
##
## loaded via a namespace (and not attached):
## [1] TH.data_1.1-2 minqa_1.2.5 colorspace_2.1-0
## [4] ggsignif_0.6.4 estimability_1.4.1 parameters_0.21.1
## [7] rstudioapi_0.15.0 ggpubr_0.6.0 farver_2.1.1
## [10] ggrepel_0.9.3 fansi_1.0.4 mvtnorm_1.2-2
## [13] mathjaxr_1.6-0 codetools_0.2-18 splines_4.2.2
## [16] knitr_1.43 polyclip_1.10-4 zeallot_0.1.0
## [19] jsonlite_1.8.7 nloptr_2.0.3 broom_1.0.5
## [22] ggforce_0.4.1 compiler_4.2.2 emmeans_1.8.7
## [25] backports_1.4.1 Matrix_1.6-0 fastmap_1.1.1
## [28] cli_3.6.1 tweenr_2.0.2 htmltools_0.5.5
## [31] tools_4.2.2 lmerTest_3.1-3 coda_0.19-4
## [34] gtable_0.3.3 glue_1.6.2 Rcpp_1.0.11
## [37] carData_3.0-5 cellranger_1.1.0 vctrs_0.6.3
## [40] nlme_3.1-160 insight_0.19.3 xfun_0.39
## [43] openxlsx_4.2.5.2 lme4_1.1-34 timechange_0.2.0
## [46] lifecycle_1.0.3 rstatix_0.7.2 dendextend_1.17.1
## [49] MASS_7.3-60 zoo_1.8-12 scales_1.2.1
## [52] ragg_1.2.5 hms_1.1.3 sandwich_3.0-2
## [55] rematch2_2.1.2 RColorBrewer_1.1-3 yaml_2.3.7
## [58] curl_5.0.1 gridExtra_2.3 reshape_0.8.9
## [61] stringi_1.7.12 paletteer_1.5.0 bayestestR_0.13.1
## [64] boot_1.3-28 zip_2.3.0 systemfonts_1.0.4
## [67] rlang_1.1.1 pkgconfig_2.0.3 evaluate_0.21
## [70] lattice_0.20-45 patchwork_1.1.2 htmlwidgets_1.6.2
## [73] labeling_0.4.2 tidyselect_1.2.0 GGally_2.1.2
## [76] plyr_1.8.8 magrittr_2.0.3 R6_2.5.1
## [79] generics_0.1.3 multcomp_1.4-25 pillar_1.9.0
## [82] haven_2.5.3 foreign_0.8-83 withr_2.5.0
## [85] survival_3.4-0 datawizard_0.8.0 abind_1.4-5
## [88] car_3.1-2 utf8_1.2.3 correlation_0.8.4
## [91] tzdb_0.4.0 rmarkdown_2.23 viridis_0.6.3
## [94] grid_4.2.2 readxl_1.4.3 data.table_1.14.8
## [97] digest_0.6.33 xtable_1.8-4 numDeriv_2016.8-1.1
## [100] textshaping_0.3.6 statsExpressions_1.5.1 munsell_0.5.0
## [103] viridisLite_0.4.2